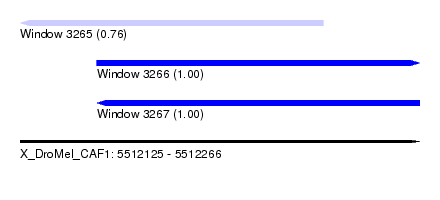
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,512,125 – 5,512,266 |
| Length | 141 |
| Max. P | 0.999220 |

| Location | 5,512,125 – 5,512,232 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.27 |
| Mean single sequence MFE | -25.15 |
| Consensus MFE | -16.33 |
| Energy contribution | -16.82 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
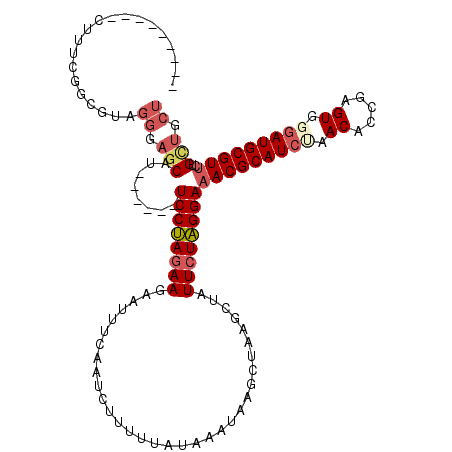
>X_DroMel_CAF1 5512125 107 - 22224390 AAGUAUUUCAAUCUUUUUAUAAAUAAGCUAAGCUAUUCUAGGAAACGCAUCAAACAACGAGUGAGAUGCGUUAUGCUGCUGUUUA----UGCAAUGGCUCU---------UCAAUUACUU (((((....................(((...))).....((((((((((((..((.....))..))))))))..(((..(((...----.)))..))))))---------)....))))) ( -20.20) >DroSec_CAF1 2252 107 - 1 AAGAAUUUCAAUCUUUUUAUAAAUAAGCUAAGCUAUUCUAGGAAACGCAUCUAACACCGAGUGGGAUGCGUUCUGUUGCUGUUUA----UGCAAUUGCUCU---------UCAAUUACUU ((((..................((((((..(((.......((((.(((((((.((.....)).)))))))))))...))))))))----)((....)))))---------)......... ( -21.00) >DroSim_CAF1 2150 102 - 1 AAGAAUUCC-AUCUUUUU-UAAAUAAGCUAAGCUAUUCUAGGA-ACGCAU-UAACACCGAGUG-GAUGCGUUCUGCUGCUGUUUA----UGCAAUGGCUCC---------UCAAUUACUU ..((...((-((......-((((((.((..(((.......(((-((((((-(.((.....)).-)))))))))))))))))))))----....))))....---------))........ ( -21.81) >DroEre_CAF1 2277 114 - 1 AA------CAACCUUUUUAUAAAUAAACUAAGCUAUUCUGGGAAACGCAUCCUACACCUCGUGGGAUGCGUUCAGCCGCUGUCUACGUUUGCAGUAGCCUUUUAAAGUAACCAGUAGCUU ..------.....................((((((..(((((((.((((((((((.....))))))))))))).((..((((........))))..))............)))))))))) ( -37.60) >consensus AAGAAUUUCAAUCUUUUUAUAAAUAAGCUAAGCUAUUCUAGGAAACGCAUCUAACACCGAGUGGGAUGCGUUCUGCUGCUGUUUA____UGCAAUGGCUCU_________UCAAUUACUU ...............................((((((...((.(((((((((.((.....)).)))))))))...))((...........))))))))...................... (-16.33 = -16.82 + 0.50)

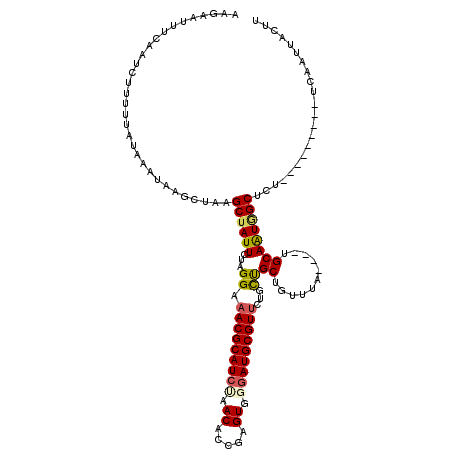
| Location | 5,512,152 – 5,512,266 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.03 |
| Mean single sequence MFE | -30.47 |
| Consensus MFE | -17.64 |
| Energy contribution | -17.89 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.58 |
| SVM decision value | 3.44 |
| SVM RNA-class probability | 0.999220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5512152 114 + 22224390 AGCAGCAUAACGCAUCUCACUCGUUGUUUGAUGCGUUUCCUAGAAUAGCUUAGCUUAUUUAUAAAAAGAUUGAAAUACUUCUAGGA------AUGCUCCCUACUGCGACAGCUCUUCAAA .((((...((((((((..((.....))..))))))))((((((((((..(((((((.........))).))))..)).))))))))------..........)))).............. ( -28.40) >DroSec_CAF1 2279 105 + 1 AGCAACAGAACGCAUCCCACUCGGUGUUAGAUGCGUUUCCUAGAAUAGCUUAGCUUAUUUAUAAAAAGAUUGAAAUUCUUCUAGGA------AUGCUCCCUACGCCGAAAG--------- .((........)).......(((((((..(..(((((.(((((((....(((((((.........))).)))).....))))))))------))))..)..)))))))...--------- ( -26.60) >DroSim_CAF1 2177 100 + 1 AGCAGCAGAACGCAUC-CACUCGGUGUUA-AUGCGU-UCCUAGAAUAGCUUAGCUUAUUUA-AAAAAGAU-GGAAUUCUUCUAGGA------AUGCUCCCUACGCCGAAAG--------- ....((.....))...-...(((((((..-..((((-((((((((...((...(((.....-...)))..-)).....))))))))------)))).....)))))))...--------- ( -31.10) >DroEre_CAF1 2317 108 + 1 AGCGGCUGAACGCAUCCCACGAGGUGUAGGAUGCGUUUCCCAGAAUAGCUUAGUUUAUUUAUAAAAAGGUUG------UUAUGGGAGCCAUUAUGAUCUGUAA------AUUGCUUGAAU (((((((((((((((((.((.....)).))))))))))((((.((((((((..((((....)))).))))))------)).)))))))).((((.....))))------...)))..... ( -35.80) >consensus AGCAGCAGAACGCAUCCCACUCGGUGUUAGAUGCGUUUCCUAGAAUAGCUUAGCUUAUUUAUAAAAAGAUUGAAAUUCUUCUAGGA______AUGCUCCCUACGCCGAAAG_________ ((((....((((((((..((.....))..))))))))((((((((...........((((......))))........)))))))).......))))....................... (-17.64 = -17.89 + 0.25)

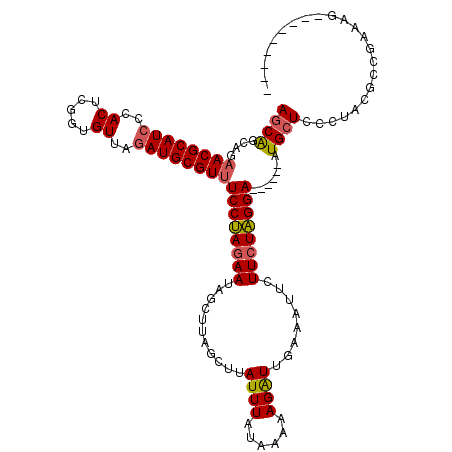

| Location | 5,512,152 – 5,512,266 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.03 |
| Mean single sequence MFE | -29.17 |
| Consensus MFE | -19.17 |
| Energy contribution | -20.86 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.998014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5512152 114 - 22224390 UUUGAAGAGCUGUCGCAGUAGGGAGCAU------UCCUAGAAGUAUUUCAAUCUUUUUAUAAAUAAGCUAAGCUAUUCUAGGAAACGCAUCAAACAACGAGUGAGAUGCGUUAUGCUGCU ..............((((((........------((((((((.(((((............)))))(((...))).))))))))((((((((..((.....))..)))))))).)))))). ( -30.00) >DroSec_CAF1 2279 105 - 1 ---------CUUUCGGCGUAGGGAGCAU------UCCUAGAAGAAUUUCAAUCUUUUUAUAAAUAAGCUAAGCUAUUCUAGGAAACGCAUCUAACACCGAGUGGGAUGCGUUCUGUUGCU ---------.....((((.(.(((((.(------(((((((((.....)................(((...))).)))))))))..((((((.((.....)).))))))))))).))))) ( -29.80) >DroSim_CAF1 2177 100 - 1 ---------CUUUCGGCGUAGGGAGCAU------UCCUAGAAGAAUUCC-AUCUUUUU-UAAAUAAGCUAAGCUAUUCUAGGA-ACGCAU-UAACACCGAGUG-GAUGCGUUCUGCUGCU ---------........(((((((((.(------((((((((.......-..(((...-.....)))........))))))))-).((((-(.((.....)).-)))))))))).)))). ( -26.83) >DroEre_CAF1 2317 108 - 1 AUUCAAGCAAU------UUACAGAUCAUAAUGGCUCCCAUAA------CAACCUUUUUAUAAAUAAACUAAGCUAUUCUGGGAAACGCAUCCUACACCUCGUGGGAUGCGUUCAGCCGCU .....(((...------...((((.....((((((.......------......................))))))))))((.((((((((((((.....))))))))))))...))))) ( -30.05) >consensus _________CUUUCGGCGUAGGGAGCAU______UCCUAGAAGAAUUUCAAUCUUUUUAUAAAUAAGCUAAGCUAUUCUAGGAAACGCAUCUAACACCGAGUGGGAUGCGUUCUGCUGCU ....................((.(((........((((((((.................................))))))))(((((((((.((.....)).)))))))))..))).)) (-19.17 = -20.86 + 1.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:02 2006