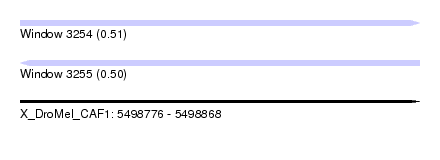
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,498,776 – 5,498,868 |
| Length | 92 |
| Max. P | 0.509792 |

| Location | 5,498,776 – 5,498,868 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 82.09 |
| Mean single sequence MFE | -43.97 |
| Consensus MFE | -27.96 |
| Energy contribution | -28.15 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5498776 92 + 22224390 ACCGGACAGAAGGCCUUUUCCGUAUUGAGGGAUUGCGGCGACAGUGCCCGGAUGGGUGCCU---UCAGUCCA-CAGCU--GCUGUCGCCGCCCUGUUG ....(((((....(((((........)))))...(((((((((((((..((((.((.....---)).)))).-..)).--))))))))))).))))). ( -37.60) >DroVir_CAF1 38018 98 + 1 ACCGGACAGAAGGCCUUAUCGGUGUUGAGGCUUUGCGGCGACAGCGCCCGGCCGUGGGCCCGUUCCAAUCGGCUGGCUGCGCUGCCGCCGCCCUGUUC ...(((((((((((((((.......)))))))))((((((.(((((((((((((((((.....))))..)))))))..)))))).)))))).)))))) ( -56.80) >DroGri_CAF1 27032 98 + 1 ACCGGACAGAAGGCCUUGUCAGUGUUGAGGCUAUGCGGCGACAGGGCCCGGCCGGGGGCCCGUUCUAAUCGGCUGGCUGCGCUGCCGCCGCCCUGUUC ...((((((..(((((((.......)))))))..((((((.(((.(((((((((((((....))))...)))))))..)).))).)))))).)))))) ( -47.90) >DroSim_CAF1 16801 92 + 1 ACCGGACAGAAGGCCUUUUCCGUGUUGAGGGAUUGCGGCGACAGUGCCCGGAUGGGUGCCU---UCAGUCCA-CAGCU--GCUGUCGCCGCCCUGUUG ....(((((....(((((........)))))...(((((((((((((..((((.((.....---)).)))).-..)).--))))))))))).))))). ( -37.60) >DroYak_CAF1 25197 92 + 1 ACCGGACAGAAGGCCUUCUCCGUAUUGAGGGAUUGCGGCGACAGUGCCCGGUUGGGUACCU---UGAGUCCA-CAGCU--GCUGUCGCCGCCCUGUUG ....(((((....(((((........)))))...(((((((((((....(((((((.((..---...)))).-)))))--))))))))))).))))). ( -39.90) >DroMoj_CAF1 31712 98 + 1 ACCGGACAGAAGGCCUUCUCGGUGUUGAGGCUCUGCGGCGACAGCGCCCGGCCGUGGGCCCGUUCCAAUCGGCCAGCUACGCUAUCGCCGCCCUGUUC ...((((((.((((((.(........))))).))(((((((.((((.(.(((((((((.....))))..))))).)...)))).))))))).)))))) ( -44.00) >consensus ACCGGACAGAAGGCCUUCUCCGUGUUGAGGCAUUGCGGCGACAGUGCCCGGCCGGGGGCCC___CCAAUCCA_CAGCU__GCUGUCGCCGCCCUGUUC ....(((((....((((.........))))....(((((((((((....(((((((.............))..)))))..))))))))))).))))). (-27.96 = -28.15 + 0.19)

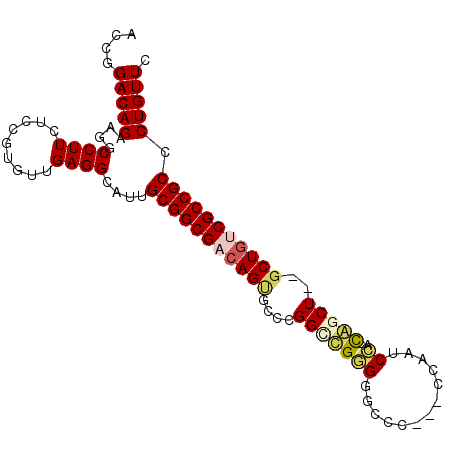
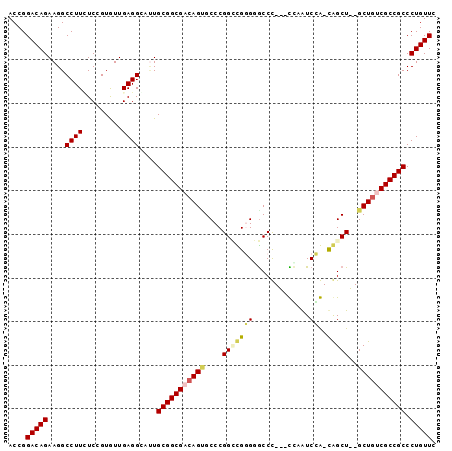
| Location | 5,498,776 – 5,498,868 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 82.09 |
| Mean single sequence MFE | -40.02 |
| Consensus MFE | -28.15 |
| Energy contribution | -27.23 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5498776 92 - 22224390 CAACAGGGCGGCGACAGC--AGCUG-UGGACUGA---AGGCACCCAUCCGGGCACUGUCGCCGCAAUCCCUCAAUACGGAAAAGGCCUUCUGUCCGGU .....((((((((((((.--.(((.-((((.((.---.......))))))))).))))))))))...........((((((......))))))))... ( -33.80) >DroVir_CAF1 38018 98 - 1 GAACAGGGCGGCGGCAGCGCAGCCAGCCGAUUGGAACGGGCCCACGGCCGGGCGCUGUCGCCGCAAAGCCUCAACACCGAUAAGGCCUUCUGUCCGGU (.((((((((((((((((((..((.((((..(((.......))))))).)))))))))))))))...((((...........))))..))))).)... ( -48.70) >DroGri_CAF1 27032 98 - 1 GAACAGGGCGGCGGCAGCGCAGCCAGCCGAUUAGAACGGGCCCCCGGCCGGGCCCUGUCGCCGCAUAGCCUCAACACUGACAAGGCCUUCUGUCCGGU (.((((((((((((((((.......))..........((((((......)))))))))))))))...((((...........))))..))))).)... ( -40.60) >DroSim_CAF1 16801 92 - 1 CAACAGGGCGGCGACAGC--AGCUG-UGGACUGA---AGGCACCCAUCCGGGCACUGUCGCCGCAAUCCCUCAACACGGAAAAGGCCUUCUGUCCGGU .....((((((((((((.--.(((.-((((.((.---.......))))))))).))))))))))...........((((((......))))))))... ( -33.90) >DroYak_CAF1 25197 92 - 1 CAACAGGGCGGCGACAGC--AGCUG-UGGACUCA---AGGUACCCAACCGGGCACUGUCGCCGCAAUCCCUCAAUACGGAGAAGGCCUUCUGUCCGGU ..(((((((((((((((.--.(((.-((....))---.))).(((....)))..)))))))))).....(((......))).......)))))..... ( -31.80) >DroMoj_CAF1 31712 98 - 1 GAACAGGGCGGCGAUAGCGUAGCUGGCCGAUUGGAACGGGCCCACGGCCGGGCGCUGUCGCCGCAGAGCCUCAACACCGAGAAGGCCUUCUGUCCGGU (.((((((((((((((((((..(((((((..(((.......)))))))))))))))))))))))((.(((((........).))))))))))).)... ( -51.30) >consensus CAACAGGGCGGCGACAGC__AGCUG_CCGACUGA___AGGCACCCAGCCGGGCACUGUCGCCGCAAACCCUCAACACGGAAAAGGCCUUCUGUCCGGU ..(((((((((((((((....(((..(......)....(((.....))).))).))))))))))....(((...........)))...)))))..... (-28.15 = -27.23 + -0.91)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:51 2006