| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,475,930 – 5,476,030 |
| Length | 100 |
| Max. P | 0.628831 |

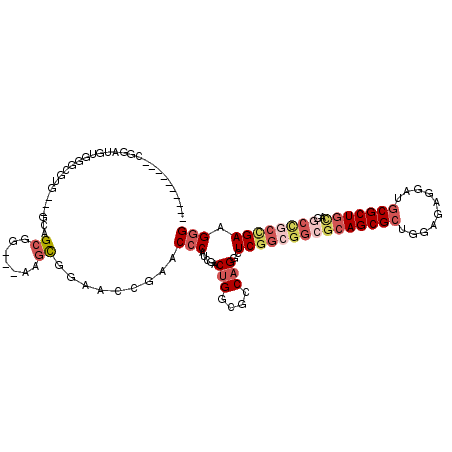
| Location | 5,475,930 – 5,476,030 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 74.42 |
| Mean single sequence MFE | -48.27 |
| Consensus MFE | -26.00 |
| Energy contribution | -27.87 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.628831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
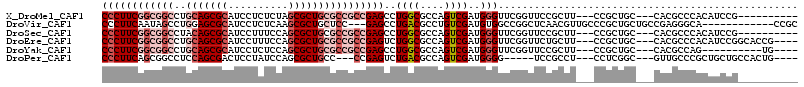
>X_DroMel_CAF1 5475930 100 + 22224390 ----------CGGAUGUGGGCGUG---GCAGCGG---AAGCGGAACCGAACCCAUCGACUGGCGCCAGGCUCGGCGGCGCAGCGCUAGAGAGGAUGCGCUGCAGGCCGCCGAAGGG ----------.......((((.((---((.((..---..))((........))..........)))).))))(((((((((((((..........)))))))..))))))...... ( -49.30) >DroVir_CAF1 9251 101 + 1 GCGG------------UGCCCUCGGCAGCAGCGGGCAACGUUGAGCCGGCCACAUCGACAGGCGUCAGGCUC---GGAGCAGCGCUUGAGAGGAUGCGCUCCAGGCUAUUGAAGGG ....------------..((((((((..(((((.....))))).))))(((.........))).((((((..---(((((.(((..(....)..))))))))..)))..))))))) ( -39.10) >DroSec_CAF1 1290 100 + 1 ----------CGGAUGUGGGCGUG---GCAGCGG---AAGCGGAACCGAACCCAUCGACUGGCGCCAGGCUCGGCGGCGCAGCGCUGGAAAGGAUGCGCUGUAGGCCGCCGAAGGG ----------.......((((.((---((.((..---..))((........))..........)))).))))(((((((((((((..........)))))))..))))))...... ( -47.90) >DroEre_CAF1 10031 106 + 1 ----CGGUGCCGGAUGUGGGCGUG---GCAGCGG---AAGCAGAACCGAACCCAUCGACUGGCGCCAGACUCGGCGGCGCAGCGCUGGAAAGGAUGCGCUGCAGGCCGCCGAAGGG ----.((((((((.((((((..((---(..((..---..))....)))..)))).)).))))))))....(((((((((((((((..........)))))))..)))))))).... ( -61.40) >DroYak_CAF1 1727 96 + 1 ----CA----------CUGGCGUG---GCAGCGG---AAGCGGAACCGAACCCAUCGACUGGCGCCAGGCUCGGCGGCGCAGCGCUGGAGAGGAUGCGCUGCAGGCCGCCGAAGGG ----..----------(((((((.---...((..---..))((........))........)))))))..(((((((((((((((..........)))))))..)))))))).... ( -50.20) >DroPer_CAF1 1668 98 + 1 ----CAGUGGCAGCAGCGGGCAAC---GCCGAGG---AGGCGGA-----CCCCAUCGACUGGCGUCAGACUCGG---GGCAGCGCUGGAUAGGAGUCGCUGGAGGCCGCUGAAGGG ----(((((((..(((((((...(---(((....---.))))..-----.)))...(((....))).(((((.(---..((....))..)..)))))))))...)))))))..... ( -41.70) >consensus __________CGGAUGUGGGCGUG___GCAGCGG___AAGCGGAACCGAACCCAUCGACUGGCGCCAGGCUCGGCGGCGCAGCGCUGGAGAGGAUGCGCUGCAGGCCGCCGAAGGG ..............................((.......)).........(((.....(((....)))..(((((((((((((((..........)))))))..)))))))).))) (-26.00 = -27.87 + 1.86)


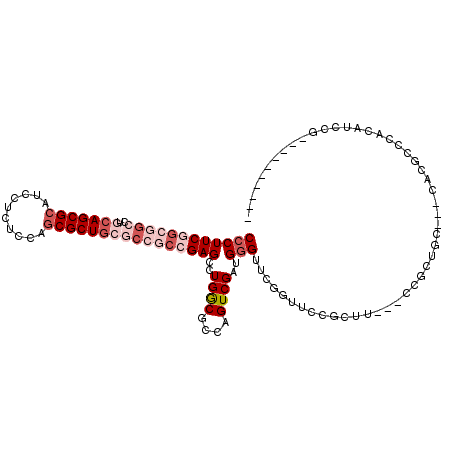
| Location | 5,475,930 – 5,476,030 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 74.42 |
| Mean single sequence MFE | -42.75 |
| Consensus MFE | -24.14 |
| Energy contribution | -26.42 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.56 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5475930 100 - 22224390 CCCUUCGGCGGCCUGCAGCGCAUCCUCUCUAGCGCUGCGCCGCCGAGCCUGGCGCCAGUCGAUGGGUUCGGUUCCGCUU---CCGCUGC---CACGCCCACAUCCG---------- ....((((((((..(((((((..........)))))))))))))))(.(((....))).)(.(((((..(((..((...---.))..))---)..)))))).....---------- ( -44.60) >DroVir_CAF1 9251 101 - 1 CCCUUCAAUAGCCUGGAGCGCAUCCUCUCAAGCGCUGCUCC---GAGCCUGACGCCUGUCGAUGUGGCCGGCUCAACGUUGCCCGCUGCUGCCGAGGGCA------------CCGC ....(((...((.((((((((............)).)))))---).)).))).((((.(((..(..((.(((........))).))..)...))))))).------------.... ( -34.30) >DroSec_CAF1 1290 100 - 1 CCCUUCGGCGGCCUACAGCGCAUCCUUUCCAGCGCUGCGCCGCCGAGCCUGGCGCCAGUCGAUGGGUUCGGUUCCGCUU---CCGCUGC---CACGCCCACAUCCG---------- ....((((((((...((((((..........)))))).))))))))(.(((....))).)(.(((((..(((..((...---.))..))---)..)))))).....---------- ( -41.30) >DroEre_CAF1 10031 106 - 1 CCCUUCGGCGGCCUGCAGCGCAUCCUUUCCAGCGCUGCGCCGCCGAGUCUGGCGCCAGUCGAUGGGUUCGGUUCUGCUU---CCGCUGC---CACGCCCACAUCCGGCACCG---- ...(((((((((..(((((((..........))))))))))))))))...((.(((.(..(.(((((..(((...((..---..)).))---)..))))))..).))).)).---- ( -52.30) >DroYak_CAF1 1727 96 - 1 CCCUUCGGCGGCCUGCAGCGCAUCCUCUCCAGCGCUGCGCCGCCGAGCCUGGCGCCAGUCGAUGGGUUCGGUUCCGCUU---CCGCUGC---CACGCCAG----------UG---- ...(((((((((..(((((((..........)))))))))))))))).((((((.((((....(((..((....))..)---)))))).---..))))))----------..---- ( -48.10) >DroPer_CAF1 1668 98 - 1 CCCUUCAGCGGCCUCCAGCGACUCCUAUCCAGCGCUGCC---CCGAGUCUGACGCCAGUCGAUGGGG-----UCCGCCU---CCUCGGC---GUUGCCCGCUGCUGCCACUG---- .....(((.(((...(((((.........((((((((..---..(((.(.(((.(((.....))).)-----)).).))---)..))))---))))..)))))..))).)))---- ( -35.90) >consensus CCCUUCGGCGGCCUGCAGCGCAUCCUCUCCAGCGCUGCGCCGCCGAGCCUGGCGCCAGUCGAUGGGUUCGGUUCCGCUU___CCGCUGC___CACGCCCACAUCCG__________ ((((((((((((..(((((((..........))))))))))))))))..((((....))))..))).................................................. (-24.14 = -26.42 + 2.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:45 2006