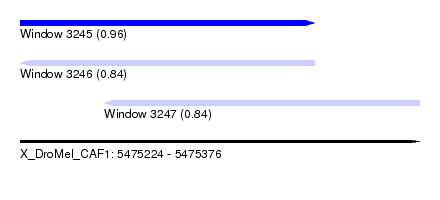
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,475,224 – 5,475,376 |
| Length | 152 |
| Max. P | 0.963584 |

| Location | 5,475,224 – 5,475,336 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 73.79 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -23.84 |
| Energy contribution | -24.32 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5475224 112 + 22224390 GUCCUGCUCCCGACAC--CCGUCACUCCUCGUCCAGACCUGAUGAAUCCCACAAUAAUGGCCAGAAUGGCGAACGCCAUCAGGAUGCAGUGGCUGGCGUCGGCGAGGUAAAUGG ......((((((((.(--(.((((((...(((((..............(((......)))...((.((((....)))))).))))).)))))).)).))))).)))........ ( -39.50) >DroPse_CAF1 979 84 + 1 ------------------------------AACCAGACCUGAUGAAGCCCACUAUGAUGGCCAGUAUGGCGAAGGCCAUGAGGAUGCAGUGGCUGGCAUCGGCCAGGUAAAUGG ------------------------------..(((.(((((.(((.(((..((((.....((..((((((....)))))).)).....))))..))).)))..)))))...))) ( -34.30) >DroYak_CAF1 1022 113 + 1 GU-CUGCUCCCGACACUCCCCGCACUCCUCGUCCAGACCUGAUGAAUCCCACAAUAAUGGCCAGUAUGGCGAAGGCCAUCAGGAUGCAGUGGCUGGCGUCGGCGAGGUAAAUGG ..-...((((((((.((..((((.....((.....))((((((.....(((......))).......(((....))))))))).....))))..)).))))).)))........ ( -36.50) >DroMoj_CAF1 1895 93 + 1 -------------------CG--CCUUCUGUUACGCACCUUAUGAAGCCCACUAUGAUGGCCAGUAUGGCAAAGGCCAUAAGAAUGCAGUGACUGGCAUCGGCCAGGUAAAUGG -------------------.(--((....((((((((.((((((..(((..(......)(((.....)))...)))))))))..))).)))))((((....)))))))...... ( -27.80) >DroAna_CAF1 1024 108 + 1 AU-CAGCUCCCAGU-C--CCAUC--CCAGUAUCCAGACCUGAUAAAGCCCACUAUUAUGGCCAGUAUGGCGAAGGCCAUCAGGAUGCAAUGACUGGCAUCGGCCAGGUAGAUGG ..-...........-.--(((((--...((((((.(.(((((((........))))).)).).(.(((((....)))))).)))))).....(((((....)))))...))))) ( -35.20) >DroPer_CAF1 983 84 + 1 ------------------------------AACCAGACCUGAUGAAGCCCACUAUGAUGGCCAGUAUGGCGAAGGCCAUGAGGAUGCAGUGGCUGGCAUCGGCCAGGUAAAUGG ------------------------------..(((.(((((.(((.(((..((((.....((..((((((....)))))).)).....))))..))).)))..)))))...))) ( -34.30) >consensus ___________________C_____UCCU_AUCCAGACCUGAUGAAGCCCACUAUGAUGGCCAGUAUGGCGAAGGCCAUCAGGAUGCAGUGGCUGGCAUCGGCCAGGUAAAUGG ................................(((.((((..((.....)).......((((...(((((....)))))...(((((........)))))))))))))...))) (-23.84 = -24.32 + 0.47)



| Location | 5,475,224 – 5,475,336 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 73.79 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -20.95 |
| Energy contribution | -21.48 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5475224 112 - 22224390 CCAUUUACCUCGCCGACGCCAGCCACUGCAUCCUGAUGGCGUUCGCCAUUCUGGCCAUUAUUGUGGGAUUCAUCAGGUCUGGACGAGGAGUGACGG--GUGUCGGGAGCAGGAC .......((((.((((((((.(.((((.(..((((((((.(((((((.....)))(((....)))))))))))))))((.....))).)))).).)--)))))))..).))).. ( -42.50) >DroPse_CAF1 979 84 - 1 CCAUUUACCUGGCCGAUGCCAGCCACUGCAUCCUCAUGGCCUUCGCCAUACUGGCCAUCAUAGUGGGCUUCAUCAGGUCUGGUU------------------------------ ......(((.((((((((..((((((((.......((((((...........))))))..)))).)))).)))).)))).))).------------------------------ ( -28.10) >DroYak_CAF1 1022 113 - 1 CCAUUUACCUCGCCGACGCCAGCCACUGCAUCCUGAUGGCCUUCGCCAUACUGGCCAUUAUUGUGGGAUUCAUCAGGUCUGGACGAGGAGUGCGGGGAGUGUCGGGAGCAG-AC ........(((.(((((((...((.((((((((((((((((...........))))))))((((.(((((.....))))).).)))))).))))))).))))))))))...-.. ( -43.40) >DroMoj_CAF1 1895 93 - 1 CCAUUUACCUGGCCGAUGCCAGUCACUGCAUUCUUAUGGCCUUUGCCAUACUGGCCAUCAUAGUGGGCUUCAUAAGGUGCGUAACAGAAGG--CG------------------- (((((....((((((((((........)))))..((((((....))))))..)))))....)))))((((......((.....))...)))--).------------------- ( -26.80) >DroAna_CAF1 1024 108 - 1 CCAUCUACCUGGCCGAUGCCAGUCAUUGCAUCCUGAUGGCCUUCGCCAUACUGGCCAUAAUAGUGGGCUUUAUCAGGUCUGGAUACUGG--GAUGG--G-ACUGGGAGCUG-AU .((.((.((..(((.((.(((((.(((....((((((((((...(((.....)))(((....)))))))..))))))....))))))))--.)).)--)-.)..)))).))-.. ( -40.50) >DroPer_CAF1 983 84 - 1 CCAUUUACCUGGCCGAUGCCAGCCACUGCAUCCUCAUGGCCUUCGCCAUACUGGCCAUCAUAGUGGGCUUCAUCAGGUCUGGUU------------------------------ ......(((.((((((((..((((((((.......((((((...........))))))..)))).)))).)))).)))).))).------------------------------ ( -28.10) >consensus CCAUUUACCUGGCCGAUGCCAGCCACUGCAUCCUGAUGGCCUUCGCCAUACUGGCCAUCAUAGUGGGCUUCAUCAGGUCUGGAU_AGGA_____G___________________ ((((.....((((((((((........)))))...(((((....)))))...))))).....))))................................................ (-20.95 = -21.48 + 0.53)


| Location | 5,475,256 – 5,475,376 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.83 |
| Mean single sequence MFE | -38.42 |
| Consensus MFE | -31.90 |
| Energy contribution | -31.68 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5475256 120 - 22224390 UUCUUCGUCUUGGUGCGCCAUCAGCAGUUCUCGCUGUUGGCCAUUUACCUCGCCGACGCCAGCCACUGCAUCCUGAUGGCGUUCGCCAUUCUGGCCAUUAUUGUGGGAUUCAUCAGGUCU ......(.((((((((((((((((((((.((.((.((((((..........)))))))).))..)))))....))))))))((((((.....)))(((....))))))..))))))).). ( -41.70) >DroVir_CAF1 8569 120 - 1 UUCUUCGUCUUGGUGCGUCAUCAGCAGUUCUCGCUGUUGGCCAUUUACCUGGCCGAUGCCAGUCACUGCAUUCUAAUGGCCUUUGCCAUACUCGCUAUCAUUAUCGGUUUUAUCAGGUGC ....(((...(((((((......(((((.((.((.((((((((......)))))))))).))..)))))......(((((....)))))...))).))))....)))............. ( -35.30) >DroGri_CAF1 1382 120 - 1 UUCUUCGUCUUGGUGCGUCAUCAGCAGUUCUCGCUGUUGGCCAUUUACCUCGCCGAUGCCAGUCACUGUAUUCUAAUGGCCUUUGCCAUACUAGCCAUCAUUAUUGGCUUCAUCAGGUGC ...........((((.((..((((((((....))))))))..)).)))).((((((((.................(((((....)))))...(((((.......))))).)))).)))). ( -32.10) >DroWil_CAF1 23008 120 - 1 UUCUUCGUCUUGGUGCGUCAUCAGAAGUUCUCGCUAUUGGCCGUUUAUCUGGCCGAUGCCAGCCAUUGUAUACUGAUGGCAUUUGCCAUUUUGGCCAUCAUAGUGGGCUUUAUAAGGUAA ..((((....(((((...)))))))))..((((((((((((((......((((.(((((((..((........)).))))))).))))...))))))..))))))))............. ( -37.40) >DroMoj_CAF1 1908 120 - 1 UUCUUCGUCUUGGUGCGUCAUCAGCAGUUCUCGCUGUUGGCCAUUUACCUGGCCGAUGCCAGUCACUGCAUUCUUAUGGCCUUUGCCAUACUGGCCAUCAUAGUGGGCUUCAUAAGGUGC ......(((((((((.((((...(((((.((.((.((((((((......)))))))))).))..))))).....((((((....)))))).)))))))))....))))............ ( -40.20) >DroAna_CAF1 1052 120 - 1 UUCUUCGUCUUGGUGCGCCAUCAGCAGUUCUCGCUGUUGGCCAUCUACCUGGCCGAUGCCAGUCAUUGCAUCCUGAUGGCCUUCGCCAUACUGGCCAUAAUAGUGGGCUUUAUCAGGUCU ......(.(((((((.((((((((((((.((.((.((((((((......)))))))))).))..)))))....)))))))....(((.(((((.......))))))))..))))))).). ( -43.80) >consensus UUCUUCGUCUUGGUGCGUCAUCAGCAGUUCUCGCUGUUGGCCAUUUACCUGGCCGAUGCCAGUCACUGCAUUCUGAUGGCCUUUGCCAUACUGGCCAUCAUAGUGGGCUUCAUCAGGUGC ...(((((..(((((.((((...(((((.((.((.((((((((......)))))))))).))..)))))......(((((....)))))..)))))))))..)))))............. (-31.90 = -31.68 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:44 2006