| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,463,661 – 5,463,758 |
| Length | 97 |
| Max. P | 0.800520 |

| Location | 5,463,661 – 5,463,758 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 75.50 |
| Mean single sequence MFE | -23.40 |
| Consensus MFE | -17.57 |
| Energy contribution | -17.18 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.800520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5463661 97 - 22224390 UGACUAACCAUC----CCCUUACCCAU-----UUACCCGCCCACGCACAGUGUACAAACUGCAGCUGGAGGCACCGGCUCCAUGUCCACUGCGCUGCCAGCGAACA ............----...........-----.....(((.((.((.(((((.(((...((.((((((.....)))))).))))).))))).))))...))).... ( -25.00) >DroSec_CAF1 3472 97 - 1 UGACUAACCAUC----CCUUUACCCAU-----UUAACCGCCUCCGCACAGUGUACAAACUGCAGCUGGAGGCACCGGCUCCAUGUCCACUGCGCUGCCAGCGAACA ............----...........-----.....(((..(.((.(((((.(((...((.((((((.....)))))).))))).))))).)).)...))).... ( -24.60) >DroSim_CAF1 3213 97 - 1 UGACUAACCAUC----CCCUUACCCAU-----UUACCCGCCCCCGUAUAGUGUACAAACUGCAGCUGGAGGCACCGGCUCCAUGUCCACUGCGCUGCCAGCGAACA ............----...........-----.....(((..(.(..(((((.(((...((.((((((.....)))))).))))).)))))..).)...))).... ( -21.00) >DroEre_CAF1 2460 106 - 1 UGAUUCACCAUCAUCCCCCCUCCCCCUCCUCCUCCACCACCACCACACAGUGUACAAACUGCAGCUGGAGGCACCGGCUCCAUGUCCACUGCGCUGCCAGCGAACA ((((.....))))..................................(((((.(((...((.((((((.....)))))).))))).)))))(((.....))).... ( -20.60) >DroYak_CAF1 2513 88 - 1 UGAUUAACCAUCAUCCCCCUUCCCC------------------CGCACAGUGUACAAGCUGCAGCUGGAGGCACCGGCUCCAUGUCCAUUGCGCUGCCAGCGAACA ((((.....))))............------------------(((.(((((((...((((.((((((.....)))))).)).))....)))))))...))).... ( -21.80) >DroPer_CAF1 1444 88 - 1 UCUCUCUCUCUC-------UCUCUU--------GGCA---CUCUCUCUAGUGUACAAGAUCCAGCUGGAGGCCCCGGCCCCGUGUCCGAUGCGGUGCCAGCGCACC ............-------..((((--------((((---((......))))).)))))....(((((.(((....)))((((((...))))))..)))))..... ( -27.40) >consensus UGACUAACCAUC____CCCUUACCCAU_____UUACCCGCCCCCGCACAGUGUACAAACUGCAGCUGGAGGCACCGGCUCCAUGUCCACUGCGCUGCCAGCGAACA ............................................((.(((((.(((...((.((((((.....)))))).))))).))))).))............ (-17.57 = -17.18 + -0.38)


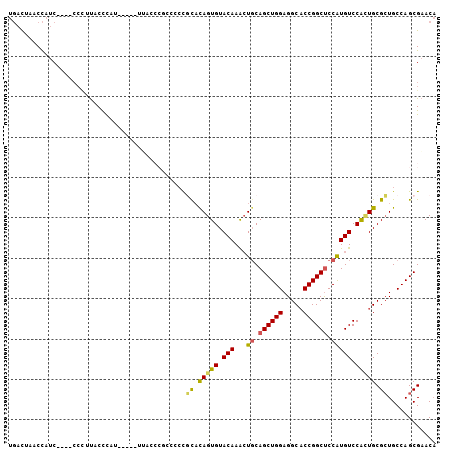
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:39 2006