
| Sequence ID | X_DroMel_CAF1 |
|---|---|
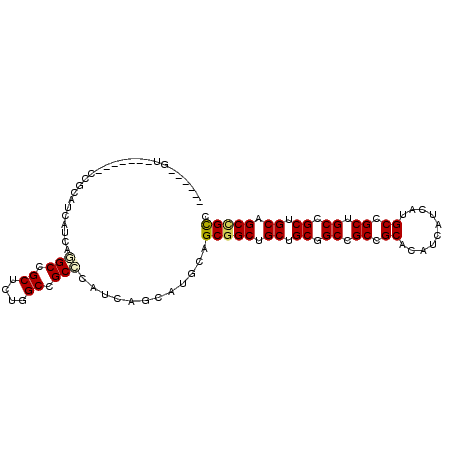
| Location | 5,451,630 – 5,451,728 |
| Length | 98 |
| Max. P | 0.993377 |

| Location | 5,451,630 – 5,451,728 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 77.40 |
| Mean single sequence MFE | -47.57 |
| Consensus MFE | -27.43 |
| Energy contribution | -27.52 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
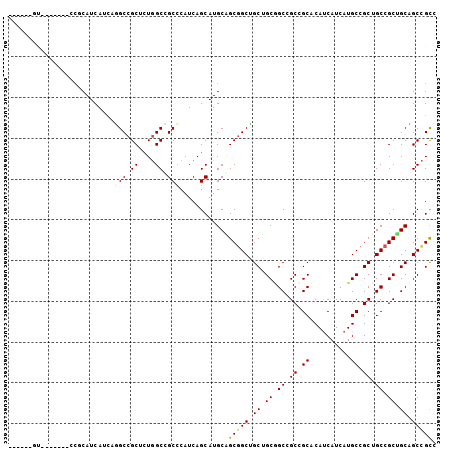
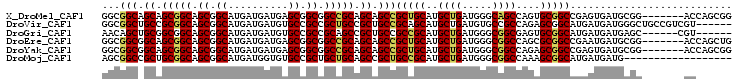
>X_DroMel_CAF1 5451630 98 + 22224390 CCGCUGGU-------CCGCAUCACUCGGCCGCACUGGCUGCCCAUCAGCAUGCAGCGGCUGCUGCGGCCGCCGCUCAUCAUCAUGCCGCUGCCGCUGCUGCCGCC ..((((((-------..(((.......(((.....))))))..)))))).....(((((.((.(((((.((.((..........)).)).))))).)).))))). ( -43.01) >DroVir_CAF1 135722 99 + 1 ------ACGACGGCAGCCCAUCAUCAUGCCGCUCUGGCGGCACAUCAGCAUGCUGCGGCAGCGGCAGCGGCGGCACAUCAUCAUGCCGCUGCCGCGGCAGCCGCC ------....((((.(((........((((((...((((((......)).))))))))))((((((((((((...........))))))))))))))).)))).. ( -55.10) >DroGri_CAF1 113735 93 + 1 ------ACG------GCUCAUCAUCAUGCCGCACUCGCCGCCCAUCAGCAUGCGGCGGCAGCGGCUGCGGCGGCACAUCAUCAUGCCGCUGCCGCCGCAGCUGUU ------.((------((..........)))).....(((((((((....))).))))))(((((((((((((((((((....)))....)))))))))))))))) ( -51.80) >DroEre_CAF1 70205 98 + 1 CAGCUGGU-------CCGCAUCAUUCGGCCGCGCUGGCCGCCCAUCAGCAUGCAGCGGCUGCUGCGGCCGCCGCUCAUCAUCAUGCCGCUGCCGCUGCCGCCGCC ..((((((-------..((.......((((.....))))))..)))))).....(((((.((.(((((.((.((..........)).)).))))).)).))))). ( -44.61) >DroYak_CAF1 60107 98 + 1 CCGCUGGU-------CCGCAUCACUCGGCCGCUCUGGCCGCCCAUCAGCAUGCAGCGGCUGCUGCGGCCGCCGCUCAUCAUCAUGCCGCUGCCGCUGCCGCCGCC ..((((((-------..((.......((((.....))))))..)))))).....(((((.((.(((((.((.((..........)).)).))))).)).))))). ( -44.71) >DroMoj_CAF1 141976 87 + 1 ------------------CAUCAUCAUGCCGCUUUGGCCGCCCAUCAGCAUGCGGCAGCGGCUGCAGCAGCGGCACACCAUCAUGCCGCUGCCGCAGCGGCCGCU ------------------........((((((....((.........))..))))))((((((((.(((((((((........)))))))))....)))))))). ( -46.20) >consensus ______GU_______CCGCAUCAUCAGGCCGCUCUGGCCGCCCAUCAGCAUGCAGCGGCUGCUGCGGCCGCCGCACAUCAUCAUGCCGCUGCCGCUGCAGCCGCC ..........................(((.((....)).)))............(((((.((.((.((.((.((..........)).)).)).)).)).))))). (-27.43 = -27.52 + 0.08)

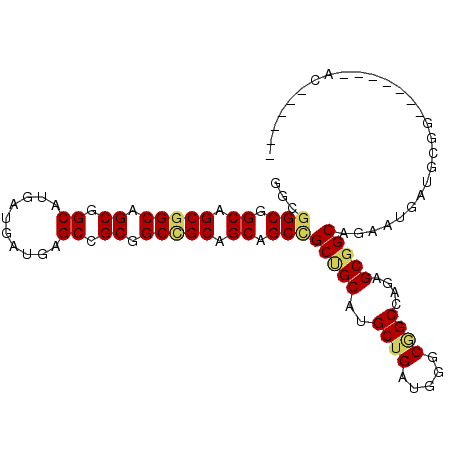
| Location | 5,451,630 – 5,451,728 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 77.40 |
| Mean single sequence MFE | -51.80 |
| Consensus MFE | -33.25 |
| Energy contribution | -33.37 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5451630 98 - 22224390 GGCGGCAGCAGCGGCAGCGGCAUGAUGAUGAGCGGCGGCCGCAGCAGCCGCUGCAUGCUGAUGGGCAGCCAGUGCGGCCGAGUGAUGCGG-------ACCAGCGG (((.((..(((((((((((((.((.((.((.((....)))))).)))))))))).)))))....)).)))..(((((((((....).)))-------.)).))). ( -46.50) >DroVir_CAF1 135722 99 - 1 GGCGGCUGCCGCGGCAGCGGCAUGAUGAUGUGCCGCCGCUGCCGCUGCCGCAGCAUGCUGAUGUGCCGCCAGAGCGGCAUGAUGAUGGGCUGCCGUCGU------ ((((((.(((((((((((((((((.((......)).)).))))))))))))..(((.(...((((((((....))))))))..))))))).))))))..------ ( -59.00) >DroGri_CAF1 113735 93 - 1 AACAGCUGCGGCGGCAGCGGCAUGAUGAUGUGCCGCCGCAGCCGCUGCCGCCGCAUGCUGAUGGGCGGCGAGUGCGGCAUGAUGAUGAGC------CGU------ ..(((((((((((((((((((.......((((....))))))))))))))))))).))))....(((((..((.((......))))..))------)))------ ( -52.31) >DroEre_CAF1 70205 98 - 1 GGCGGCGGCAGCGGCAGCGGCAUGAUGAUGAGCGGCGGCCGCAGCAGCCGCUGCAUGCUGAUGGGCGGCCAGCGCGGCCGAAUGAUGCGG-------ACCAGCUG (((.((..(((((((((((((.((.((.((.((....)))))).)))))))))).)))))....)).)))(((..(((((.......)))-------.)).))). ( -48.50) >DroYak_CAF1 60107 98 - 1 GGCGGCGGCAGCGGCAGCGGCAUGAUGAUGAGCGGCGGCCGCAGCAGCCGCUGCAUGCUGAUGGGCGGCCAGAGCGGCCGAGUGAUGCGG-------ACCAGCGG .((((((((.(((((.((.((..........)).)).)))))....)))))))).(((((.....(((((.....))))).(.....)..-------..))))). ( -47.60) >DroMoj_CAF1 141976 87 - 1 AGCGGCCGCUGCGGCAGCGGCAUGAUGGUGUGCCGCUGCUGCAGCCGCUGCCGCAUGCUGAUGGGCGGCCAAAGCGGCAUGAUGAUG------------------ .(((((.((((((((((((((((......)))))))))))))))).)))))(((((((((.(((....)))...))))))).))...------------------ ( -56.90) >consensus GGCGGCGGCAGCGGCAGCGGCAUGAUGAUGAGCCGCGGCCGCAGCAGCCGCUGCAUGCUGAUGGGCGGCCAGAGCGGCAGAAUGAUGCGG_______AC______ ...(((.((.(((((.((.((..........)).)).))))).)).)))(((((..((((.....))))....)))))........................... (-33.25 = -33.37 + 0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:34 2006