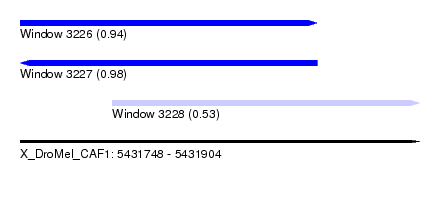
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,431,748 – 5,431,904 |
| Length | 156 |
| Max. P | 0.975142 |

| Location | 5,431,748 – 5,431,864 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 89.22 |
| Mean single sequence MFE | -22.73 |
| Consensus MFE | -22.02 |
| Energy contribution | -21.80 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
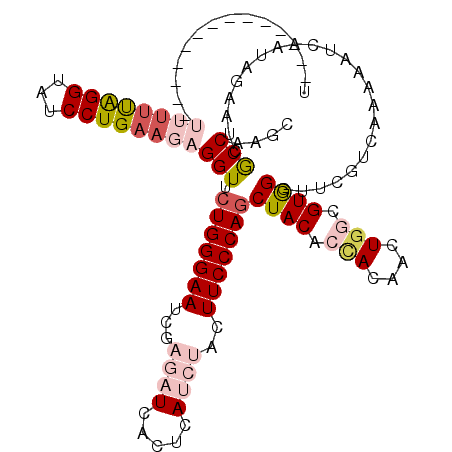
>X_DroMel_CAF1 5431748 116 + 22224390 UUUGCCCAGCAUCCUCUUAUUUUUUAGCGGCUCCUUUUUUCUUCUUUUUUUUUUUUGGGUAUCCUGAAGAGGUUCUGGGAAUCGAGAUCACUCAUCUACUUCCCAGCUACACGACA .......(((..(((((......((((.((..(((.....................)))..)))))))))))...((((((...((((.....))))..)))))))))........ ( -21.80) >DroSec_CAF1 38751 102 + 1 UUUGCCCAGCAUCCUCUUUUUUCUUAGCGGCUCCUUU--------------UUUUUAGGUAUCCUGAAGAGGUUCUGGGAAUCGAGAUCACUCAUCUACUUCCCAGCUACACUACA .......(((..(((((......((((.((..(((..--------------.....)))..)))))))))))...((((((...((((.....))))..)))))))))........ ( -23.20) >DroSim_CAF1 38445 102 + 1 UUUGCCCAGCAUCCUCUUUUUUCUUAGCGGCUCCUUU--------------UUUUUAGGUAUCCUGAAGAGGUUCUGGGAAUCGAGAUCACUCAUCUACUUCCCAGCUACACUACA .......(((..(((((......((((.((..(((..--------------.....)))..)))))))))))...((((((...((((.....))))..)))))))))........ ( -23.20) >consensus UUUGCCCAGCAUCCUCUUUUUUCUUAGCGGCUCCUUU______________UUUUUAGGUAUCCUGAAGAGGUUCUGGGAAUCGAGAUCACUCAUCUACUUCCCAGCUACACUACA .......(((..(((((......((((.((..(((.....................)))..)))))))))))...((((((...((((.....))))..)))))))))........ (-22.02 = -21.80 + -0.22)



| Location | 5,431,748 – 5,431,864 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 89.22 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -26.23 |
| Energy contribution | -26.57 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.975142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5431748 116 - 22224390 UGUCGUGUAGCUGGGAAGUAGAUGAGUGAUCUCGAUUCCCAGAACCUCUUCAGGAUACCCAAAAAAAAAAAAGAAGAAAAAAGGAGCCGCUAAAAAAUAAGAGGAUGCUGGGCAAA ....((.((((((((((...((.((....))))..))))))...((((((..((....))......................(....)..........))))))..)))).))... ( -27.60) >DroSec_CAF1 38751 102 - 1 UGUAGUGUAGCUGGGAAGUAGAUGAGUGAUCUCGAUUCCCAGAACCUCUUCAGGAUACCUAAAAA--------------AAAGGAGCCGCUAAGAAAAAAGAGGAUGCUGGGCAAA ....((.((((((((((...((.((....))))..))))))...((((((..((...(((.....--------------..)))..))..........))))))..)))).))... ( -28.80) >DroSim_CAF1 38445 102 - 1 UGUAGUGUAGCUGGGAAGUAGAUGAGUGAUCUCGAUUCCCAGAACCUCUUCAGGAUACCUAAAAA--------------AAAGGAGCCGCUAAGAAAAAAGAGGAUGCUGGGCAAA ....((.((((((((((...((.((....))))..))))))...((((((..((...(((.....--------------..)))..))..........))))))..)))).))... ( -28.80) >consensus UGUAGUGUAGCUGGGAAGUAGAUGAGUGAUCUCGAUUCCCAGAACCUCUUCAGGAUACCUAAAAA______________AAAGGAGCCGCUAAGAAAAAAGAGGAUGCUGGGCAAA ....((.((((((((((...((.((....))))..))))))...((((((..((...(((.....................)))..))..........))))))..)))).))... (-26.23 = -26.57 + 0.33)


| Location | 5,431,784 – 5,431,904 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.93 |
| Mean single sequence MFE | -25.24 |
| Consensus MFE | -17.08 |
| Energy contribution | -19.42 |
| Covariance contribution | 2.34 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5431784 120 + 22224390 UUUUCUUCUUUUUUUUUUUUGGGUAUCCUGAAGAGGUUCUGGGAAUCGAGAUCACUCAUCUACUUCCCAGCUACACGACAACUGGCGUGGGUUUGUCAUAAAUCAAUAGAAUCGCCAAGC ..............(((((..((...))..)))))((.(((((((...((((.....))))..)))))))..))........(((((.....(((........)))......)))))... ( -26.50) >DroSec_CAF1 38787 106 + 1 U--------------UUUUUAGGUAUCCUGAAGAGGUUCUGGGAAUCGAGAUCACUCAUCUACUUCCCAGCUACACUACAACUGGCGUGGGUUCGCCAAAAAUCAAUAGAAUCGCCAAGC .--------------((((((((...))))))))(((((((((((...((((.....))))..)))))..............(((((......))))).........))))))....... ( -28.10) >DroSim_CAF1 38481 106 + 1 U--------------UUUUUAGGUAUCCUGAAGAGGUUCUGGGAAUCGAGAUCACUCAUCUACUUCCCAGCUACACUACAACUGGCGUGGGUUCGUCAAAAAUCAAUAGAAUCGCCAAGC (--------------((((((((...)))))))))((.(((((((...((((.....))))..)))))))..))........(((((...((((..............)))))))))... ( -27.04) >DroEre_CAF1 49379 91 + 1 -----------------UUCAGGUAUCCUGAAGAGGUUCUGGGAAUCCAGA------------UUCCCAGCUACAACAUAACUGGAGUAGGUUCGUCAAAAAUCAAUAGAAUCGCCAAGC -----------------((((((...))))))(..((.((((((((....)------------)))))))..))..).....(((....(((((..............))))).)))... ( -24.44) >DroYak_CAF1 40281 91 + 1 -----------------AUCAGGUAUCCUGCAGAGGUUCUGGGAAUCCACA------------UUCCCAACUACAACAUAACUGCUGUAGGUUCGUCAACAAUCAAUUAAAUCACCAAGC -----------------....(((..(((((((.((((.(((((((....)------------))))))..........)))).)))))))...(....).............))).... ( -20.10) >consensus U______________UUUUUAGGUAUCCUGAAGAGGUUCUGGGAAUCGAGAUCACUCAUCUACUUCCCAGCUACACCACAACUGGCGUGGGUUCGUCAAAAAUCAAUAGAAUCGCCAAGC ...............((((((((...))))))))(((.(((((((...((((.....))))..)))))))((((.(((....))).)))).......................))).... (-17.08 = -19.42 + 2.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:27 2006