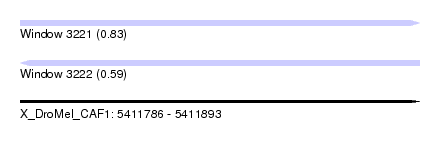
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,411,786 – 5,411,893 |
| Length | 107 |
| Max. P | 0.825109 |

| Location | 5,411,786 – 5,411,893 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 91.93 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -26.48 |
| Energy contribution | -26.35 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.825109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
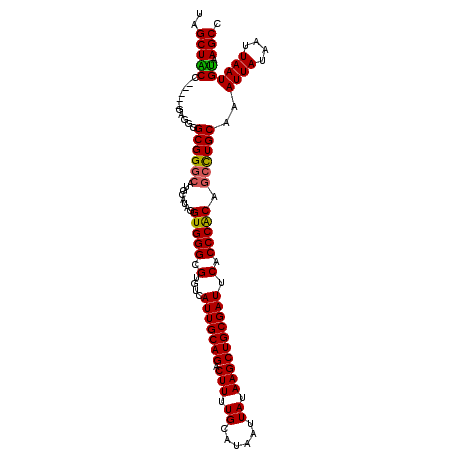
>X_DroMel_CAF1 5411786 107 + 22224390 GGCUUACAUUAAUUAUAAUUUGCAGGCGGUGGGUGAAUCGCAGCUUAUAAUUAUGCAAAAGUCUGCAAUGACACGCCCACCUAUGCAUGCCCGCCCCUC-----GGUAGCUG (((.................((((...((((((((....(((...........)))....(((......))).))))))))..))))((((........-----))))))). ( -31.40) >DroSec_CAF1 19829 107 + 1 GGCUUACAUUAAUUAUAAUUUGCAGGCUGUGGGUGAAUCGCAGCUUAUAAUUAUGCAAAAGUCUGCAAUGACACGCCCACCUAUGCAUCGCCGCCCCUC-----GGUAGCUA ((((................((((....(((((((....(((...........)))....(((......))).)))))))...))))..((((.....)-----))))))). ( -29.10) >DroEre_CAF1 27913 107 + 1 GGCUUACAUUAAUUAUAAUUUGCAGGCUGCGGGUGAAUCGCAGCUUAUAAUUAUGCAAAAGUCUGCAAUGACACGCCCACCUAUGCAUGCCCGCCCCUC-----GGCAGCCA ((((..........((((((...(((((((((.....)))))))))..))))))(((...(((......)))..((........)).)))..(((....-----))))))). ( -31.00) >DroYak_CAF1 20046 112 + 1 AGCUUGCAUUAAUUAUAAUUUGCAAGCUGCGGGUGAAUCGCAGCUUAUAAUUAUGCAAAAGUCUGCAAUGACACGCCCACCUAUGCAAGCCCGCCCCCCAGCUGGGCAGCUA .((((((((....(((((((...(((((((((.....)))))))))..))))))).....(((......)))..........))))))))..((((.......))))..... ( -38.00) >consensus GGCUUACAUUAAUUAUAAUUUGCAGGCUGCGGGUGAAUCGCAGCUUAUAAUUAUGCAAAAGUCUGCAAUGACACGCCCACCUAUGCAUGCCCGCCCCUC_____GGCAGCUA ((((................((((....(((((((....(((...........)))....(((......))).)))))))...)))).....(((.........))))))). (-26.48 = -26.35 + -0.13)



| Location | 5,411,786 – 5,411,893 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 91.93 |
| Mean single sequence MFE | -35.40 |
| Consensus MFE | -26.85 |
| Energy contribution | -26.48 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5411786 107 - 22224390 CAGCUACC-----GAGGGGCGGGCAUGCAUAGGUGGGCGUGUCAUUGCAGACUUUUGCAUAAUUAUAAGCUGCGAUUCACCCACCGCCUGCAAAUUAUAAUUAAUGUAAGCC ..(((.(.-----...)))).(((.((((..((((((.(....(((((((.(((.((......)).)))))))))).).))))))...))))..(((((.....)))))))) ( -32.20) >DroSec_CAF1 19829 107 - 1 UAGCUACC-----GAGGGGCGGCGAUGCAUAGGUGGGCGUGUCAUUGCAGACUUUUGCAUAAUUAUAAGCUGCGAUUCACCCACAGCCUGCAAAUUAUAAUUAAUGUAAGCC ..(((.(.-----...))))(((..((((..(((.((.(((..(((((((.(((.((......)).)))))))))).)))))...)))))))..(((((.....)))))))) ( -29.00) >DroEre_CAF1 27913 107 - 1 UGGCUGCC-----GAGGGGCGGGCAUGCAUAGGUGGGCGUGUCAUUGCAGACUUUUGCAUAAUUAUAAGCUGCGAUUCACCCGCAGCCUGCAAAUUAUAAUUAAUGUAAGCC .((((((.-----((((.((.(((((((........)))))))...))...)))).)).(((((((((((((((.......)))))).......))))))))).....)))) ( -35.41) >DroYak_CAF1 20046 112 - 1 UAGCUGCCCAGCUGGGGGGCGGGCUUGCAUAGGUGGGCGUGUCAUUGCAGACUUUUGCAUAAUUAUAAGCUGCGAUUCACCCGCAGCUUGCAAAUUAUAAUUAAUGCAAGCU ..(((.(((....))).))).(((((((((..((..(((.(((......)))...)))((((((.(((((((((.......)))))))))..)))))).))..))))))))) ( -45.00) >consensus UAGCUACC_____GAGGGGCGGGCAUGCAUAGGUGGGCGUGUCAUUGCAGACUUUUGCAUAAUUAUAAGCUGCGAUUCACCCACAGCCUGCAAAUUAUAAUUAAUGUAAGCC ..(((((...........((((((........(((((.(....(((((((.(((.((......)).)))))))))).).))))).))))))..((((....)))))).))). (-26.85 = -26.48 + -0.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:22 2006