
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,394,068 – 5,394,190 |
| Length | 122 |
| Max. P | 0.911487 |

| Location | 5,394,068 – 5,394,168 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 83.13 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -15.80 |
| Energy contribution | -15.80 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5394068 100 + 22224390 ---UG--------CGUGG---CA---AGUGAGUUUUAUGAAAUUUAUGCGCAUUUCACUAGAAAAUUUAAUUAGGAAAUCUAAUUUGCUCAAAAGCGCAUAAAAAAUGUUGUCGCUG- ---..--------.((((---((---(((((...))))....(((((((((.((((....))))....((((((.....)))))).........))))))))).....)))))))..- ( -22.10) >DroVir_CAF1 35288 102 + 1 -------------AAUGGCAUGA---AGUGAAUUUUAUGAAAUUUAUGCGCAUUUCACUAGAAAAUUUAAUUAGGAAAUCUAAUUUGCUCAAAAGCGCAUAAAAAAUGUCGCCCAACG -------------..((((((((---((....)))))))...(((((((((.((((....))))....((((((.....)))))).........))))))))).........)))... ( -20.90) >DroGri_CAF1 33607 100 + 1 ---------------GAGGACGA---AGUGAAUUUUAUGAAAUUUAUGCGCAUUUCACUAGAAAAUUUAAUUAGGAAAUCUAAUUUGCUCAAAAGCGCAUAAAAAAUGUCGCCCAACG ---------------..((.(((---.((((...))))....(((((((((.((((....))))....((((((.....)))))).........))))))))).....))).)).... ( -19.30) >DroWil_CAF1 40114 111 + 1 ---UGGCUCUGGCUCUGG---CUCUUGGGGGAUUUUAUGAAAUUUAUGCGCAUUUCACUAGAAAAUUUAAUUAGGAAAUCUAAUUUGCUCAAAAGCGCAUAAAAAAUGUUGUCGCUC- ---.((((........))---))...(((.(((..(((....(((((((((.((((....))))....((((((.....)))))).........)))))))))..)))..))).)))- ( -24.10) >DroMoj_CAF1 42613 102 + 1 -------------AACAGCAUGA---AGUGAAUUUUAUGAAAUUUAUGCGCAUUUCACUAGAAAAUUUAAUUAGGAAAUCUAAUUUGCUCAAAAGCGCAUAAAAAAUGUCGCCCAGCG -------------....((.((.---.(((((((((.......((((((((.((((....))))....((((((.....)))))).........))))))))))))).)))).)))). ( -19.31) >DroAna_CAF1 19949 106 + 1 UCCAG-----AGUCCUGG---CG---AGUGAAUUUUAUGAAAUUUAUGCGCAUUUCACUAGAAAAUUUAAUUAGGAAAUCUAAUUUGCUCAAAAGCGCAUAAAAAAUGUUGUCGCUC- .((((-----....))))---.(---(((((....(((....(((((((((.((((....))))....((((((.....)))))).........)))))))))..)))...))))))- ( -26.70) >consensus _____________AAUGG___CA___AGUGAAUUUUAUGAAAUUUAUGCGCAUUUCACUAGAAAAUUUAAUUAGGAAAUCUAAUUUGCUCAAAAGCGCAUAAAAAAUGUCGCCCAUC_ ..........................................(((((((((.((((....))))....((((((.....)))))).........)))))))))............... (-15.80 = -15.80 + -0.00)



| Location | 5,394,068 – 5,394,168 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 83.13 |
| Mean single sequence MFE | -19.80 |
| Consensus MFE | -16.00 |
| Energy contribution | -16.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.505274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5394068 100 - 22224390 -CAGCGACAACAUUUUUUAUGCGCUUUUGAGCAAAUUAGAUUUCCUAAUUAAAUUUUCUAGUGAAAUGCGCAUAAAUUUCAUAAAACUCACU---UG---CCACG--------CA--- -..(((.........(((((((((.........((((((.....))))))....((((....)))).)))))))))..........(.....---.)---...))--------).--- ( -18.20) >DroVir_CAF1 35288 102 - 1 CGUUGGGCGACAUUUUUUAUGCGCUUUUGAGCAAAUUAGAUUUCCUAAUUAAAUUUUCUAGUGAAAUGCGCAUAAAUUUCAUAAAAUUCACU---UCAUGCCAUU------------- .....((((......(((((((((.........((((((.....))))))....((((....)))).)))))))))(((....)))......---...))))...------------- ( -18.60) >DroGri_CAF1 33607 100 - 1 CGUUGGGCGACAUUUUUUAUGCGCUUUUGAGCAAAUUAGAUUUCCUAAUUAAAUUUUCUAGUGAAAUGCGCAUAAAUUUCAUAAAAUUCACU---UCGUCCUC--------------- ....((((((.....(((((((((.........((((((.....))))))....((((....)))).)))))))))................---))))))..--------------- ( -22.00) >DroWil_CAF1 40114 111 - 1 -GAGCGACAACAUUUUUUAUGCGCUUUUGAGCAAAUUAGAUUUCCUAAUUAAAUUUUCUAGUGAAAUGCGCAUAAAUUUCAUAAAAUCCCCCAAGAG---CCAGAGCCAGAGCCA--- -(.((..........(((((((((.........((((((.....))))))....((((....)))).)))))))))..................(.(---(....)))...))).--- ( -18.10) >DroMoj_CAF1 42613 102 - 1 CGCUGGGCGACAUUUUUUAUGCGCUUUUGAGCAAAUUAGAUUUCCUAAUUAAAUUUUCUAGUGAAAUGCGCAUAAAUUUCAUAAAAUUCACU---UCAUGCUGUU------------- ....(((((.(((.....))))))))...((((((((((.....))))))..........(((((((........)))))))..........---...))))...------------- ( -18.20) >DroAna_CAF1 19949 106 - 1 -GAGCGACAACAUUUUUUAUGCGCUUUUGAGCAAAUUAGAUUUCCUAAUUAAAUUUUCUAGUGAAAUGCGCAUAAAUUUCAUAAAAUUCACU---CG---CCAGGACU-----CUGGA -(((.((........(((((((((.........((((((.....))))))....((((....)))).)))))))))...........)).))---).---((((....-----)))). ( -23.71) >consensus _GAGCGACAACAUUUUUUAUGCGCUUUUGAGCAAAUUAGAUUUCCUAAUUAAAUUUUCUAGUGAAAUGCGCAUAAAUUUCAUAAAAUUCACU___UC___CCAGG_____________ ...............(((((((((.........((((((.....))))))....((((....)))).))))))))).......................................... (-16.00 = -16.00 + 0.00)

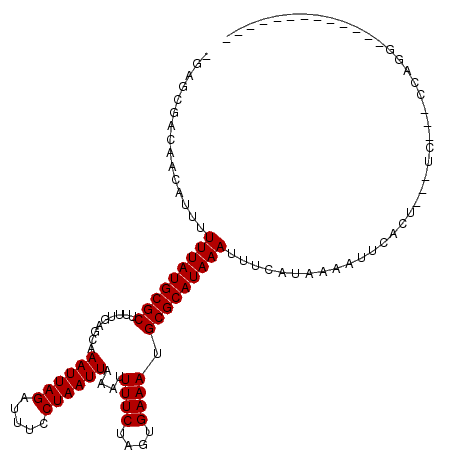
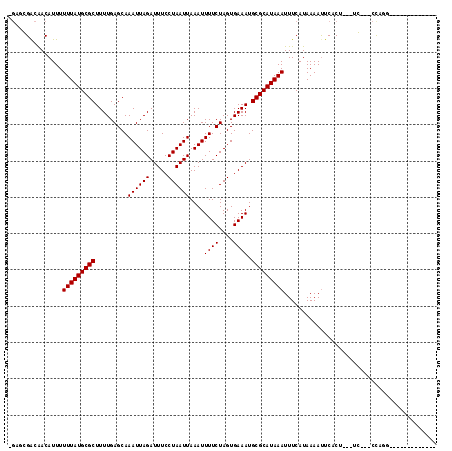
| Location | 5,394,090 – 5,394,190 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.39 |
| Mean single sequence MFE | -20.47 |
| Consensus MFE | -15.80 |
| Energy contribution | -15.80 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5394090 100 + 22224390 AAAUUUAUGCGCAUUUCACUAGAAAAUUUAAUUAGGAAAUCUAAUUUGCUCAAAAGCGCAUAAAAAAUGUUGUCGCUGCUAAAGCG-----GAU--GGGGUUU-GUGU--- ...(((((((((.((((....))))....((((((.....)))))).........)))))))))((((.(..(((((.....))).-----)).--.).))))-....--- ( -24.70) >DroPse_CAF1 22634 95 + 1 AAAUUUAUGCGCAUUUCACUAGAAAAUUUAAUUAGGAAAUCUAAUUUGCUCAAAAGCGCAUAAAAAAUGUUGUCGCUCUGG----G-----UCC--UGGGUUC-GUG---- ...(((((((((.((((....))))....((((((.....)))))).........)))))))))..........((((.(.----.-----..)--.))))..-...---- ( -18.70) >DroEre_CAF1 10834 104 + 1 AAAUUUAUGCGCAUUUCACUAGAAAAUUUAAUUAGGAAAUCUAAUUUGCUCAAAAGCGCAUAAAAAAUGUUGUCGCUUCUGAAGGG-----GAA--GGCGGAGGGUGAGGG ...(((((((((.((((....))))....((((((.....)))))).........)))))))))......(.(((((((((.....-----...--..)))).))))).). ( -22.60) >DroWil_CAF1 40147 89 + 1 AAAUUUAUGCGCAUUUCACUAGAAAAUUUAAUUAGGAAAUCUAAUUUGCUCAAAAGCGCAUAAAAAAUGUUGUCGCUCCUU----G-----UUU--UUU--------U--- ...(((((((((.((((....))))....((((((.....)))))).........))))))))).................----.-----...--...--------.--- ( -15.80) >DroYak_CAF1 2053 111 + 1 AAAUUUAUGCGCAUUUCACUAGAAAAUUUAAUUAGGAAAUCUAAUUUGCUCAAAAGCGCAUAAAAAAUGUUGUCGCUUCCAAAAGGUGGGAGUAGGGGGGAAGGGUGGGGA ...(((((((((.((((....))))....((((((.....)))))).........)))))))))...(.((.(((((((((.....))))))).)).)).).......... ( -24.30) >DroAna_CAF1 19977 83 + 1 AAAUUUAUGCGCAUUUCACUAGAAAAUUUAAUUAGGAAAUCUAAUUUGCUCAAAAGCGCAUAAAAAAUGUUGUCGCUCCUGCC---------------------------- ...(((((((((.((((....))))....((((((.....)))))).........)))))))))..........((....)).---------------------------- ( -16.70) >consensus AAAUUUAUGCGCAUUUCACUAGAAAAUUUAAUUAGGAAAUCUAAUUUGCUCAAAAGCGCAUAAAAAAUGUUGUCGCUCCUGAA__G_____GAA__GGGG____GUG____ ...(((((((((.((((....))))....((((((.....)))))).........)))))))))............................................... (-15.80 = -15.80 + -0.00)

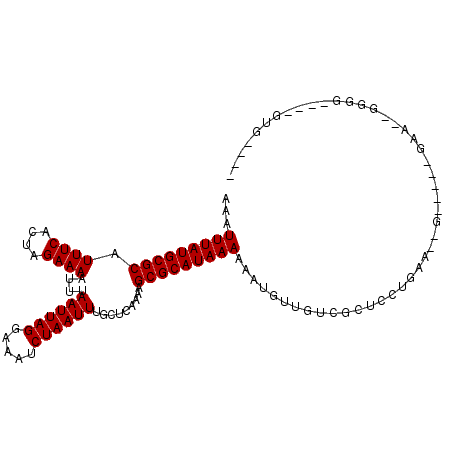
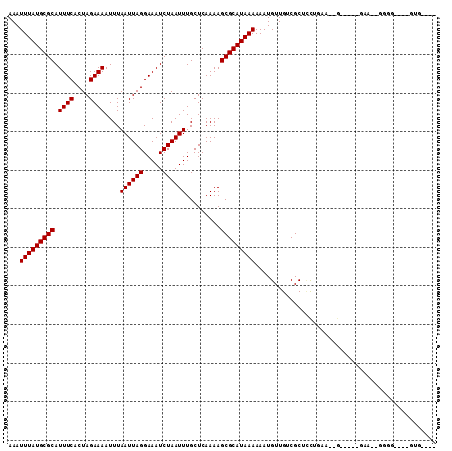
| Location | 5,394,090 – 5,394,190 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.39 |
| Mean single sequence MFE | -18.06 |
| Consensus MFE | -16.00 |
| Energy contribution | -16.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5394090 100 - 22224390 ---ACAC-AAACCCC--AUC-----CGCUUUAGCAGCGACAACAUUUUUUAUGCGCUUUUGAGCAAAUUAGAUUUCCUAAUUAAAUUUUCUAGUGAAAUGCGCAUAAAUUU ---....-.......--...-----((((.....)))).........(((((((((.........((((((.....))))))....((((....)))).)))))))))... ( -19.80) >DroPse_CAF1 22634 95 - 1 ----CAC-GAACCCA--GGA-----C----CCAGAGCGACAACAUUUUUUAUGCGCUUUUGAGCAAAUUAGAUUUCCUAAUUAAAUUUUCUAGUGAAAUGCGCAUAAAUUU ----...-.......--...-----.----.................(((((((((.........((((((.....))))))....((((....)))).)))))))))... ( -16.00) >DroEre_CAF1 10834 104 - 1 CCCUCACCCUCCGCC--UUC-----CCCUUCAGAAGCGACAACAUUUUUUAUGCGCUUUUGAGCAAAUUAGAUUUCCUAAUUAAAUUUUCUAGUGAAAUGCGCAUAAAUUU ............((.--...-----....(((((((((.((..........)))))))))))(((((((((.....))))))....((((....)))))))))........ ( -20.10) >DroWil_CAF1 40147 89 - 1 ---A--------AAA--AAA-----C----AAGGAGCGACAACAUUUUUUAUGCGCUUUUGAGCAAAUUAGAUUUCCUAAUUAAAUUUUCUAGUGAAAUGCGCAUAAAUUU ---.--------...--...-----.----.................(((((((((.........((((((.....))))))....((((....)))).)))))))))... ( -16.00) >DroYak_CAF1 2053 111 - 1 UCCCCACCCUUCCCCCCUACUCCCACCUUUUGGAAGCGACAACAUUUUUUAUGCGCUUUUGAGCAAAUUAGAUUUCCUAAUUAAAUUUUCUAGUGAAAUGCGCAUAAAUUU ......(.(((((..................))))).).........(((((((((.........((((((.....))))))....((((....)))).)))))))))... ( -19.47) >DroAna_CAF1 19977 83 - 1 ----------------------------GGCAGGAGCGACAACAUUUUUUAUGCGCUUUUGAGCAAAUUAGAUUUCCUAAUUAAAUUUUCUAGUGAAAUGCGCAUAAAUUU ----------------------------.((....))..........(((((((((.........((((((.....))))))....((((....)))).)))))))))... ( -17.00) >consensus ____CAC____CCCC__AAC_____C__UUCAGGAGCGACAACAUUUUUUAUGCGCUUUUGAGCAAAUUAGAUUUCCUAAUUAAAUUUUCUAGUGAAAUGCGCAUAAAUUU ...............................................(((((((((.........((((((.....))))))....((((....)))).)))))))))... (-16.00 = -16.00 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:18 2006