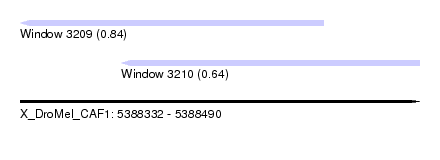
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,388,332 – 5,388,490 |
| Length | 158 |
| Max. P | 0.840225 |

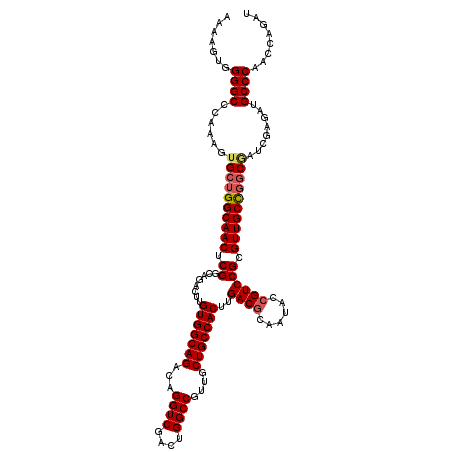
| Location | 5,388,332 – 5,388,452 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -48.41 |
| Consensus MFE | -45.23 |
| Energy contribution | -45.91 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5388332 120 - 22224390 AAAAGUGGGGCCCAAAGUGCUGGCAACUCGGCAGACUUGUGGCAGACAGGUCGACUGGCCGUUGCUGCCACUUGACGCAAUACAGUCCGCGUUGCCGGCAAUCGAGAUGCCCAGCCAGAU ....((.((((......((((((((((.((........(((((((...((((....))))....)))))))..(((........))))).))))))))))........)))).))..... ( -49.94) >DroSec_CAF1 69509 120 - 1 AAAAGUGGGGCCCAAAGUGCUGGCAACUCGGCAGACUUGUGGCAGACAGGUCGACUGGCCGUUGCUGCCACUUGACGCAAUACCGUCCGCGUUGCCGGCGAUCGAGAUGCCCAACCAGAU ....((.((((......((((((((((.((........(((((((...((((....))))....)))))))..((((......)))))).))))))))))........)))).))..... ( -50.04) >DroSim_CAF1 57221 120 - 1 AAAAGUGGGGCCCAAAGUGCUGGCAACUCGGCAGACUUGUGGCAGACAGGUCGACUGGCCGUUGCUGCCACUUGACGCAGUACCGUCCGCGUUGCUGGCGAUCGAGAUGCCCAACCAGAU ....((.((((......(((..(((((.((........(((((((...((((....))))....)))))))..((((......)))))).)))))..)))........)))).))..... ( -47.44) >DroYak_CAF1 62998 120 - 1 AAAUGAGGGGCCCAAAGAGCAGGCAACUCGGCAGACUUGUGGCAGACAGGUCGACUGGCCGAUGCUGCCACUUGACGCGAUACCGUCCGCGUUGCCGCCGUCUGAGAUGCCCAACCAGAU .......((((........(((((....(((((..(..(((((((...((((....))))....)))))))..)(((((........))))))))))..)))))....))))........ ( -46.20) >consensus AAAAGUGGGGCCCAAAGUGCUGGCAACUCGGCAGACUUGUGGCAGACAGGUCGACUGGCCGUUGCUGCCACUUGACGCAAUACCGUCCGCGUUGCCGGCGAUCGAGAUGCCCAACCAGAU .......((((......((((((((((.((........(((((((...((((....))))....)))))))..((((......)))))).))))))))))........))))........ (-45.23 = -45.91 + 0.69)


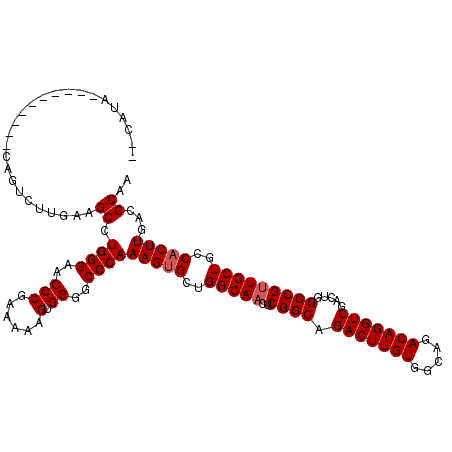
| Location | 5,388,372 – 5,388,490 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.89 |
| Mean single sequence MFE | -42.05 |
| Consensus MFE | -35.90 |
| Energy contribution | -36.40 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5388372 118 - 22224390 --CAGACGAAAUCGACCAGGCUUGCAGCCUGGGAACCCGAAAAAGUGGGGCCCAAAGUGCUGGCAACUCGGCAGACUUGUGGCAGACAGGUCGACUGGCCGUUGCUGCCACUUGACGCAA --....((....)).((((((.....))))))...((((......))))((...(((((..((((((((((..(((((((.....)))))))..))))..))))))..)))))...)).. ( -46.30) >DroSec_CAF1 69549 108 - 1 --CAUA----------CAGUCUUGAAGCCUGGGAACCCGAAAAAGUGGGGCCCAAAGUGCUGGCAACUCGGCAGACUUGUGGCAGACAGGUCGACUGGCCGUUGCUGCCACUUGACGCAA --....----------..(((........((((..(((......).))..))))(((((..((((((((((..(((((((.....)))))))..))))..))))))..)))))))).... ( -39.70) >DroSim_CAF1 57261 108 - 1 --CAUA----------CAGUCUUGAAGCCUGGGAACCCGAAAAAGUGGGGCCCAAAGUGCUGGCAACUCGGCAGACUUGUGGCAGACAGGUCGACUGGCCGUUGCUGCCACUUGACGCAG --....----------..(((........((((..(((......).))..))))(((((..((((((((((..(((((((.....)))))))..))))..))))))..)))))))).... ( -39.70) >DroYak_CAF1 63038 119 - 1 UACAUAUAAGAUCG-CCGCAUUUCAAGCCUGGGAACCCGAAAAUGAGGGGCCCAAAGAGCAGGCAACUCGGCAGACUUGUGGCAGACAGGUCGACUGGCCGAUGCUGCCACUUGACGCGA ..............-.(((...(((((..((((..(((........))).)))).......((((.((((((.(((((((.....))))))).....))))).).)))).))))).))). ( -42.50) >consensus __CAUA__________CAGUCUUGAAGCCUGGGAACCCGAAAAAGUGGGGCCCAAAGUGCUGGCAACUCGGCAGACUUGUGGCAGACAGGUCGACUGGCCGUUGCUGCCACUUGACGCAA ..........................((.((((..(((......).))..))))(((((..(((((..((((.(((((((.....))))))).....)))))))))..)))))...)).. (-35.90 = -36.40 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:12 2006