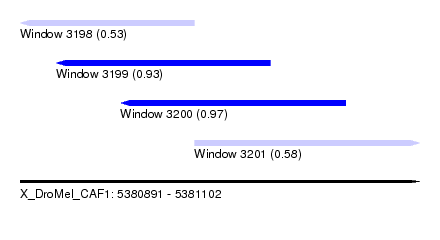
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,380,891 – 5,381,102 |
| Length | 211 |
| Max. P | 0.971483 |

| Location | 5,380,891 – 5,380,983 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 91.30 |
| Mean single sequence MFE | -21.09 |
| Consensus MFE | -17.57 |
| Energy contribution | -18.24 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.533273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5380891 92 - 22224390 UAGAGACAAAGGCGCUGAAAAGCGUGGAAAACUGGGAAAAAACUCAAAGGAUUUCGCGUAUACUCUGCCAAUAAAUAGCGCAAAUAUGAACC ..........((((((....)))((((((..((..((......))..))..)))))).........)))....................... ( -19.60) >DroSec_CAF1 62210 92 - 1 UAGAGACAAAGGCGCUGAAAAGCGGUAAAAACUGGGGAAAAACUCAAAGGACUCCGCGGAUACUCUGCCAAUAUAUAGCGCAAAUAUUGAAU ...........((((((....((((.......((((......)))).......))))((........))......))))))........... ( -21.84) >DroSim_CAF1 49168 92 - 1 UAGAGACAAAGGCGCUGAAAAGCGGUGAAAACUGGGGAAAAACUCAAAGGACUCCGCGGAUACUCUGCCAAUAUAUAGCGCAAAUAUUGAAU ...........((((((....((((.......((((......)))).......))))((........))......))))))........... ( -21.84) >consensus UAGAGACAAAGGCGCUGAAAAGCGGUGAAAACUGGGGAAAAACUCAAAGGACUCCGCGGAUACUCUGCCAAUAUAUAGCGCAAAUAUUGAAU ...........((((((....((((.......((((......)))).......))))((........))......))))))........... (-17.57 = -18.24 + 0.67)

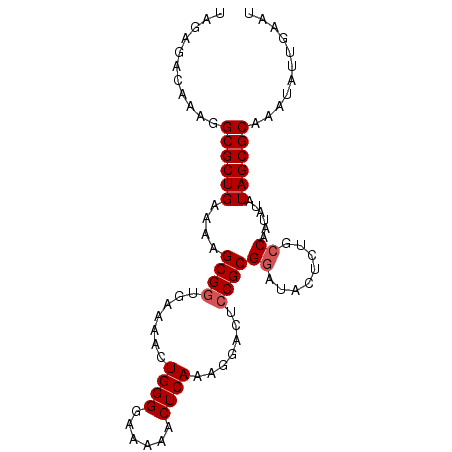
| Location | 5,380,910 – 5,381,023 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 90.90 |
| Mean single sequence MFE | -28.81 |
| Consensus MFE | -23.20 |
| Energy contribution | -23.14 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.934110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5380910 113 - 22224390 GUUGGAAAUUGAAGUUUCAGCGAAAAAAAUGGAAAGAUGCUAGAGACAA-AGGCGCUGAAAAGCGUGGAAAACUGGGAAAAAACUCAAAGGAUUUCGCGUAUACUCUGCCAAUA (((((.........((((((((.......(((.......))).......-...)))))))).((((((((..((..((......))..))..))))))))........))))). ( -27.23) >DroSec_CAF1 62229 113 - 1 GUUGGAAAUUGAAGUUCCAGCGAAAAAAGUGGAAAGAUGCUAGAGACAA-AGGCGCUGAAAAGCGGUAAAAACUGGGGAAAAACUCAAAGGACUCCGCGGAUACUCUGCCAAUA (((((((.......)))))))................((.(((((....-...((((....)))).......(((.(((....(.....)...))).)))...))))).))... ( -27.10) >DroSim_CAF1 49187 113 - 1 GUUGGAAAUUGAAGUUCCAGCGAAAAAAGCGGAAAGAUGCUAGAGACAA-AGGCGCUGAAAAGCGGUGAAAACUGGGGAAAAACUCAAAGGACUCCGCGGAUACUCUGCCAAUA (((((((.......))))))).......(((((....(((..(((....-...((((....))))((....))((((......)))).....))).))).....)))))..... ( -28.90) >DroEre_CAF1 55381 110 - 1 GU-GGAAAUUGAAGUUCCGGCGGAAAAAAUGGGAAGAUGCUAGAGACAAAAGGCGCUGAAAAGCGGGGAAAACUGGGGAAA-ACUCAAAGCACUCCUCGGAUACUCUGCCAA-- ..-((((.......))))((((((.....((((.((.((((............((((....))))........((((....-.)))).)))))).)))).....))))))..-- ( -32.00) >consensus GUUGGAAAUUGAAGUUCCAGCGAAAAAAAUGGAAAGAUGCUAGAGACAA_AGGCGCUGAAAAGCGGGGAAAACUGGGGAAAAACUCAAAGGACUCCGCGGAUACUCUGCCAAUA (((((((.......)))))))................((.(((((........((((....)))).......(((.(((..............))).)))...))))).))... (-23.20 = -23.14 + -0.06)

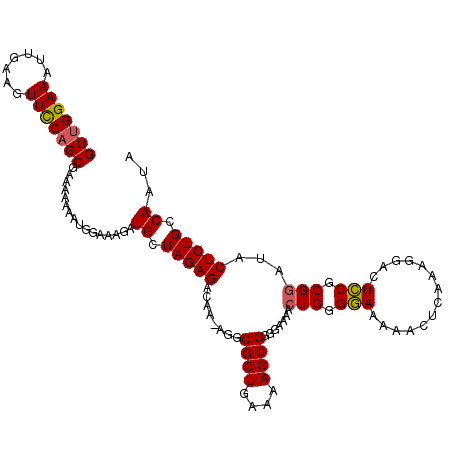

| Location | 5,380,944 – 5,381,063 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.21 |
| Mean single sequence MFE | -26.74 |
| Consensus MFE | -20.22 |
| Energy contribution | -20.82 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5380944 119 - 22224390 AAUAGCGAAAAAAGUUCGCGAGCGUUAAACUCAUAUGAAAGUUGGAAAUUGAAGUUUCAGCGAAAAAAAUGGAAAGAUGCUAGAGACAA-AGGCGCUGAAAAGCGUGGAAAACUGGGAAA ....(((((.....))))).((((((....((((......(((((((.......))))))).......))))...))))))........-..(((((....))))).............. ( -25.22) >DroSec_CAF1 62263 114 - 1 ----GCGA-AAAAGUUCGCGAGCGUUAAACUCAUAUGAAAGUUGGAAAUUGAAGUUCCAGCGAAAAAAGUGGAAAGAUGCUAGAGACAA-AGGCGCUGAAAAGCGGUAAAAACUGGGGAA ----((((-(....))))).((((((....((((......(((((((.......))))))).......))))...))))))........-...((((....))))............... ( -26.62) >DroSim_CAF1 49221 114 - 1 ----GCGA-AAAAGUUCGCGAGCGUUAAACUCAUAUGAAAGUUGGAAAUUGAAGUUCCAGCGAAAAAAGCGGAAAGAUGCUAGAGACAA-AGGCGCUGAAAAGCGGUGAAAACUGGGGAA ----((((-(....))))).((((((...(((........(((((((.......)))))))......((((......)))).)))....-.))))))......((((....))))..... ( -30.30) >DroEre_CAF1 55412 116 - 1 AAA-GCGA--AAAGUGCGCGACCGUUAAACUCAUAUGAAAGU-GGAAAUUGAAGUUCCGGCGGAAAAAAUGGGAAGAUGCUAGAGACAAAAGGCGCUGAAAAGCGGGGAAAACUGGGGAA ...-(((.--......)))..(((((...(((..........-((((.......))))((((...............)))).)))........((((....)))).....))).)).... ( -19.96) >DroYak_CAF1 54957 119 - 1 AAUGGCGA-AAAAGUUCGCGAGCGUUAAACUCAUAUGAAAGUUGGAAAUUGAAGUUCCAGCGGAAAAAAUGGGAAGAUGCCGGAGACAAAAGGCGCUGAAAAGCGAGGAAAACUGGCGAA ....((((-(....))))).((((((...(((((......(((((((.......))))))).......)))))........(....)....)))))).....((.((.....)).))... ( -31.62) >consensus AA__GCGA_AAAAGUUCGCGAGCGUUAAACUCAUAUGAAAGUUGGAAAUUGAAGUUCCAGCGAAAAAAAUGGAAAGAUGCUAGAGACAA_AGGCGCUGAAAAGCGGGGAAAACUGGGGAA ....((((.......)))).((((((....((((......(((((((.......))))))).......))))...))))))............((((....))))............... (-20.22 = -20.82 + 0.60)

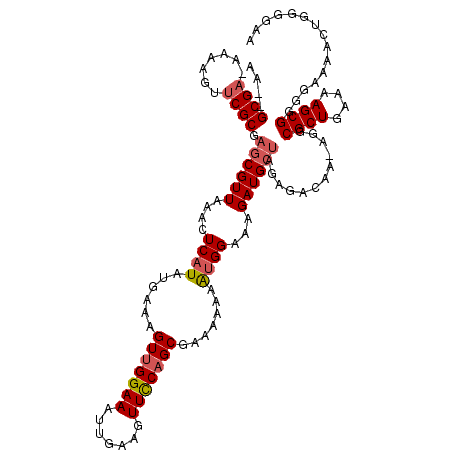
| Location | 5,380,983 – 5,381,102 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.67 |
| Mean single sequence MFE | -22.75 |
| Consensus MFE | -7.74 |
| Energy contribution | -8.74 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5380983 119 + 22224390 GCAUCUUUCCAUUUUUUUCGCUGAAACUUCAAUUUCCAACUUUCAUAUGAGUUUAACGCUCGCGAACUUUUUUCGCUAUUUCACUCGCUG-AAAAAGAAGGCUAAAAAGCUCACUGAAAA .....((((..(((((((((((((((......................((((.....))))(((((.....)))))..)))))...)).)-))))))).((((....))))....)))). ( -20.40) >DroSec_CAF1 62302 104 + 1 GCAUCUUUCCACUUUUUUCGCUGGAACUUCAAUUUCCAACUUUCAUAUGAGUUUAACGCUCGCGAACUUUU-UCGC------------UC--AAAAGAAGGCCGA-AAGCUCGCUGAAAC ((..(((((............(((((.......))))).(((((....((((.....))))(((((....)-))))------------..--....)))))..))-)))...))...... ( -21.10) >DroSim_CAF1 49260 104 + 1 GCAUCUUUCCGCUUUUUUCGCUGGAACUUCAAUUUCCAACUUUCAUAUGAGUUUAACGCUCGCGAACUUUU-UCGC------------UC--AAAAGAAGGCCGA-AAGCUCGCCGAAAC ((..(((((.((((((((...(((((.......)))))..........((((.....))))(((((....)-))))------------..--..)))))))).))-)))...))...... ( -24.30) >DroEre_CAF1 55452 105 + 1 GCAUCUUCCCAUUUUUUCCGCCGGAACUUCAAUUUCC-ACUUUCAUAUGAGUUUAACGGUCGCGCACUUU--UCGC-UUUUCGUGCGCUGGAAAAAGAAGGAUGA-AAGC---------- ...((.(((..(((((((((((((((.......))))-((((......)))).....))).((((((...--....-.....)))))).))))))))..))).))-....---------- ( -29.30) >DroYak_CAF1 54997 94 + 1 GCAUCUUCCCAUUUUUUCCGCUGGAACUUCAAUUUCCAACUUUCAUAUGAGUUUAACGCUCGCGAACUUUU-UCGCCAUUUCACUCGAUG--------------A-AAGA---------- .....................(((((.......))))).(((((((.(((((.........(((((....)-))))......))))))))--------------)-))).---------- ( -18.66) >consensus GCAUCUUUCCAUUUUUUUCGCUGGAACUUCAAUUUCCAACUUUCAUAUGAGUUUAACGCUCGCGAACUUUU_UCGC__UUUC___CG_UG__AAAAGAAGGCCGA_AAGCUC_C_GAAA_ ................((((((((((.......)))))..........((((.....)))))))))...................................................... ( -7.74 = -8.74 + 1.00)

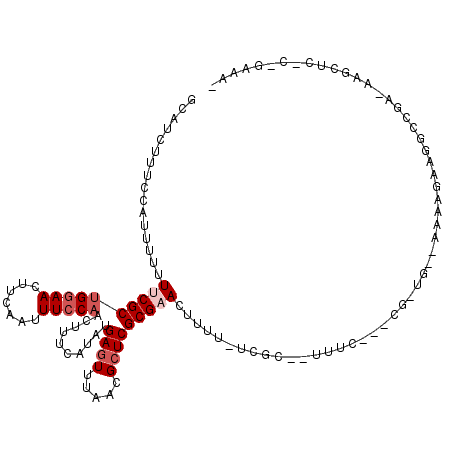

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:03 2006