
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,380,303 – 5,380,526 |
| Length | 223 |
| Max. P | 0.943690 |

| Location | 5,380,303 – 5,380,395 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.32 |
| Mean single sequence MFE | -17.80 |
| Consensus MFE | -14.48 |
| Energy contribution | -14.10 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5380303 92 - 22224390 -----------------------GCAUAUGCAAUUUGCAAUUCGCAAUUUUGCAGCCCCAGUGAGCAAAUAUUCGCCACCCACCCAAGCCAGCAAUCCGCAAACCAUAAUACCCG--- -----------------------((...(((((.((((.....))))..)))))......(((.((........)))))........))..((.....))...............--- ( -15.50) >DroSec_CAF1 61645 89 - 1 -----------------------GCAUAUGCAAUUUGCAAUUCGCAAUUUUGCAGCCCCAGUGAGCAAAUAUUCGCCACCCAC--AAGCCAGCAAUCCGCA-UCCAUAAUACCCG--- -----------------------((...(((((.((((.....))))..)))))......(((.((........)))))....--..))..((.....)).-.............--- ( -15.50) >DroSim_CAF1 48618 89 - 1 -----------------------GCAUAUGCAAUUUGCAAUUCGCAAUUUUGCAGCCCCAGUGAGCAAAUAUUCGCCACCCAC--CAGCCAGCAAUCCGCA-UCCAUAAUACCCG--- -----------------------((...(((((.((((.....))))..)))))......(((.((........)))))....--..))..((.....)).-.............--- ( -15.50) >DroYak_CAF1 54244 115 - 1 CCAUUGCCGUUGCCAUUGCCAUUGCAUAUGCAAUUUGCAAUUCGCAAUUUUGCAGCACCAGCGAGCAAAUAUUCGCCACCCAC--AAGUCCGCAAUCUGCA-CCCAUAAUACCAACAC ..(((((.(((((.(((((.((((((.........))))))..)))))...)))))....(((((......))))).......--......))))).....-................ ( -24.70) >consensus _______________________GCAUAUGCAAUUUGCAAUUCGCAAUUUUGCAGCCCCAGUGAGCAAAUAUUCGCCACCCAC__AAGCCAGCAAUCCGCA_UCCAUAAUACCCG___ .......................((...(((((.((((.....))))..)))))......(((((......)))))...........))..((.....)).................. (-14.48 = -14.10 + -0.38)


| Location | 5,380,395 – 5,380,486 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.74 |
| Mean single sequence MFE | -35.30 |
| Consensus MFE | -27.71 |
| Energy contribution | -27.75 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5380395 91 + 22224390 -------AACGGGUCGCAGGAC------UUGUCGUCCUUUUCAGGUGACAUGUGCAC----------------CUGUGCAUUCGAUUUUGCCCUGCAUUCGGGCGGGAAUUUGCCUUUGA -------..(((((((((((((------.(((((.(((....)))))))).))...)----------------))))).))))).((((((((.......))))))))............ ( -31.30) >DroSec_CAF1 61734 91 + 1 -------AACGGGUCGCAGGAC------UCGUCGUCCUUUUCAGGUGACAUGUGCAC----------------CUGUGCAUUCGAUUUUGCCCUGCAUUCGGGCGGGAAUUUGCCUUUGA -------.((((((((((((((------.....))))......(....).)))).))----------------))))(((.....((((((((.......))))))))...)))...... ( -30.60) >DroSim_CAF1 48707 91 + 1 -------AACGGGUCGCAGGAC------UCGUCGUCCUUUUCAGGUGACAUGUGCAC----------------CUGUGCAUUCGAUUUUGCCCUGCAUUCGGGCGGGAAUUUGCCUUUGA -------.((((((((((((((------.....))))......(....).)))).))----------------))))(((.....((((((((.......))))))))...)))...... ( -30.60) >DroEre_CAF1 54827 106 + 1 -------AGCGGGUCGCAGGAC------UCGUCGUCCUUUUCAGGUGACAUGUGCACCUGUGCACCUGUGCACCUGUGCAUUCGAUUUUGC-CUGCACUCGGGCGGGAAUUUGCCUUUGA -------..(((((.(((((..------..((((.......(((((((((.(((((....))))).))).))))))......))))....)-)))))))))(((((....)))))..... ( -44.82) >DroYak_CAF1 54359 104 + 1 CAACAGCAACGGGUCGCAGGAGUCGCAGUCGUCGUCCUUUUCAGGUGACAUGUGCAC----------------CUGUGCAUUCGAUUUUGCCCUGCAUUCGGGCGGGAAUUUGCCUUUGA (((..(((((((((((((((.(.((((...((((.(((....))))))).))))).)----------------))))).))))).((((((((.......))))))))..))))..))). ( -39.20) >consensus _______AACGGGUCGCAGGAC______UCGUCGUCCUUUUCAGGUGACAUGUGCAC________________CUGUGCAUUCGAUUUUGCCCUGCAUUCGGGCGGGAAUUUGCCUUUGA .........(((((.(((((..........((((.(((....)))))))..((((((..................))))))..........))))))))))(((((....)))))..... (-27.71 = -27.75 + 0.04)

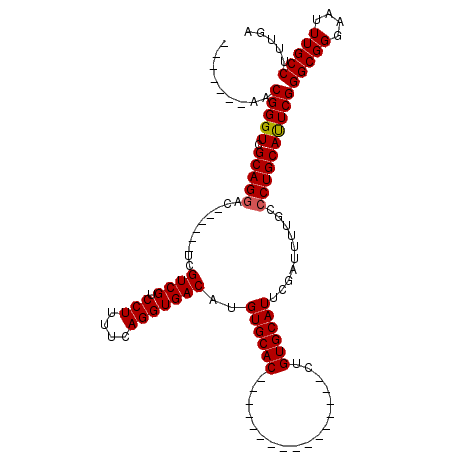
| Location | 5,380,395 – 5,380,486 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.74 |
| Mean single sequence MFE | -31.24 |
| Consensus MFE | -26.65 |
| Energy contribution | -26.89 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5380395 91 - 22224390 UCAAAGGCAAAUUCCCGCCCGAAUGCAGGGCAAAAUCGAAUGCACAG----------------GUGCACAUGUCACCUGAAAAGGACGACAA------GUCCUGCGACCCGUU------- .....(((........)))....((((((((.........((((...----------------.))))..((((.(((....)))..)))).------)))))))).......------- ( -29.40) >DroSec_CAF1 61734 91 - 1 UCAAAGGCAAAUUCCCGCCCGAAUGCAGGGCAAAAUCGAAUGCACAG----------------GUGCACAUGUCACCUGAAAAGGACGACGA------GUCCUGCGACCCGUU------- .....(((........)))....((((((((.........((((...----------------.))))..((((.(((....)))..)))).------)))))))).......------- ( -28.70) >DroSim_CAF1 48707 91 - 1 UCAAAGGCAAAUUCCCGCCCGAAUGCAGGGCAAAAUCGAAUGCACAG----------------GUGCACAUGUCACCUGAAAAGGACGACGA------GUCCUGCGACCCGUU------- .....(((........)))....((((((((.........((((...----------------.))))..((((.(((....)))..)))).------)))))))).......------- ( -28.70) >DroEre_CAF1 54827 106 - 1 UCAAAGGCAAAUUCCCGCCCGAGUGCAG-GCAAAAUCGAAUGCACAGGUGCACAGGUGCACAGGUGCACAUGUCACCUGAAAAGGACGACGA------GUCCUGCGACCCGCU------- .....(((........)))((.((((((-((.........(((((..(((((....)))))..)))))..((((.(((....)))..)))).------).))))).)).))..------- ( -38.30) >DroYak_CAF1 54359 104 - 1 UCAAAGGCAAAUUCCCGCCCGAAUGCAGGGCAAAAUCGAAUGCACAG----------------GUGCACAUGUCACCUGAAAAGGACGACGACUGCGACUCCUGCGACCCGUUGCUGUUG .(((.(((((......((((.......)))).........((((..(----------------(((((..((((.(((....)))..))))..))).)))..)))).....))))).))) ( -31.10) >consensus UCAAAGGCAAAUUCCCGCCCGAAUGCAGGGCAAAAUCGAAUGCACAG________________GUGCACAUGUCACCUGAAAAGGACGACGA______GUCCUGCGACCCGUU_______ .....(((........)))....((((((((.........(((((..................)))))..((((.(((....)))..)))).......)))))))).............. (-26.65 = -26.89 + 0.24)


| Location | 5,380,422 – 5,380,526 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.48 |
| Mean single sequence MFE | -33.46 |
| Consensus MFE | -28.81 |
| Energy contribution | -29.01 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5380422 104 + 22224390 UCAGGUGACAUGUGCAC----------------CUGUGCAUUCGAUUUUGCCCUGCAUUCGGGCGGGAAUUUGCCUUUGAGUUCGUGUGCGGCCUUUAUUGGUUUACUUCAUCAACUCGA .((((((.(....))))----------------))).(((.....((((((((.......))))))))...)))..(((((((.(((...((((......)))).....))).))))))) ( -31.20) >DroSec_CAF1 61761 104 + 1 UCAGGUGACAUGUGCAC----------------CUGUGCAUUCGAUUUUGCCCUGCAUUCGGGCGGGAAUUUGCCUUUGAGUUCGUGUGCGGCCUUUAUUGGUUUACUUCAUCAACUCGA .((((((.(....))))----------------))).(((.....((((((((.......))))))))...)))..(((((((.(((...((((......)))).....))).))))))) ( -31.20) >DroSim_CAF1 48734 104 + 1 UCAGGUGACAUGUGCAC----------------CUGUGCAUUCGAUUUUGCCCUGCAUUCGGGCGGGAAUUUGCCUUUGAGUUCGUGUGCGGCCUUUAUUGGUUUACUUCAUCAACUCGA .((((((.(....))))----------------))).(((.....((((((((.......))))))))...)))..(((((((.(((...((((......)))).....))).))))))) ( -31.20) >DroEre_CAF1 54854 119 + 1 UCAGGUGACAUGUGCACCUGUGCACCUGUGCACCUGUGCAUUCGAUUUUGC-CUGCACUCGGGCGGGAAUUUGCCUUUGAGUUCGUGUGCGGCCUUUAUUGGUUUACUUCAUCAACUCGA .(((((((((.(((((....))))).))).)))))).(((.....((((((-(((....)))))))))...)))..(((((((.(((...((((......)))).....))).))))))) ( -42.50) >DroYak_CAF1 54399 104 + 1 UCAGGUGACAUGUGCAC----------------CUGUGCAUUCGAUUUUGCCCUGCAUUCGGGCGGGAAUUUGCCUUUGAGUUCGUGUGCGGCCUUUAUUGGUUUACUUCAUCAACUCGA .((((((.(....))))----------------))).(((.....((((((((.......))))))))...)))..(((((((.(((...((((......)))).....))).))))))) ( -31.20) >consensus UCAGGUGACAUGUGCAC________________CUGUGCAUUCGAUUUUGCCCUGCAUUCGGGCGGGAAUUUGCCUUUGAGUUCGUGUGCGGCCUUUAUUGGUUUACUUCAUCAACUCGA ..(((..((..((((((..................))))))..).((((((((.......))))))))..)..)))(((((((.(((...((((......)))).....))).))))))) (-28.81 = -29.01 + 0.20)

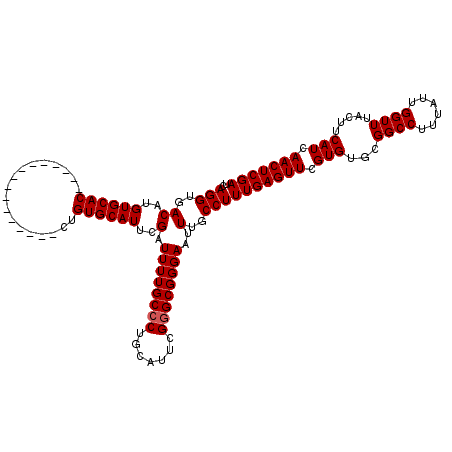
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:00 2006