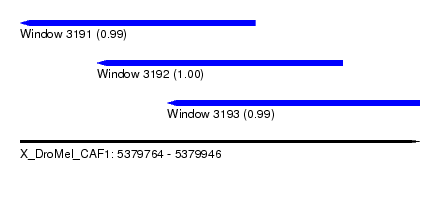
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,379,764 – 5,379,946 |
| Length | 182 |
| Max. P | 0.995539 |

| Location | 5,379,764 – 5,379,871 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.03 |
| Mean single sequence MFE | -31.09 |
| Consensus MFE | -21.20 |
| Energy contribution | -22.70 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5379764 107 - 22224390 GGACAAGGGGGCGUGGCCCCUUCGGUUGCGCCCCAUAAAAA-UGUCCUUCGAACUAUU---CACCCCUUCGUGGCCAUCCCGCCCCCUCUUCUCCCCUUUCCC-----CAUCAGGC (((.(((((((((..(((.....)))..))))))......(-((.((..((((.....---......)))).)).)))..........))).)))........-----........ ( -31.10) >DroSec_CAF1 61108 101 - 1 GGACAAGGGGGCGUGGCCCCUUCGGUUGCGCCCCAUAAAAA-UGUCCUUCGAACUAUU---UACCCCUUCCUGGCCAUCGCCC--CCU----UCCCCAAUCCC-----CGCCAGCC (((((..((((((..(((.....)))..)))))).......-)))))...........---.........(((((........--...----...........-----.))))).. ( -32.50) >DroSim_CAF1 48068 101 - 1 GGACAAGG-GGCGUGGCCCCUUCGGUUGCGCCCCAUAAAAA-UGUCCUUCGAACUAUU---CACCCCUUCCUGGCCAUCGCCCC-CCU----UCCCCAAUCCC-----CGCCAGCC (((((.((-((((..(((.....)))..)))))).......-)))))...........---.........(((((.........-...----...........-----.))))).. ( -30.96) >DroYak_CAF1 53696 107 - 1 -----AGGGGGCGUGGCCCCUUCGGUUGCGCCCCAUAAAAACUGUCCUUCGAACUACUACUCACCUCUUCCUAGACAACCCGACCACG----CCCCCACUCCCGCUGCUGCCCACC -----.((((((((((.....(((((((......................((........))...(((....)))))).)))))))))----)))))......((....))..... ( -29.80) >consensus GGACAAGGGGGCGUGGCCCCUUCGGUUGCGCCCCAUAAAAA_UGUCCUUCGAACUAUU___CACCCCUUCCUGGCCAUCCCCCC_CCU____UCCCCAAUCCC_____CGCCAGCC (((((..((((((..(((.....)))..))))))........)))))..................................................................... (-21.20 = -22.70 + 1.50)


| Location | 5,379,799 – 5,379,911 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.35 |
| Mean single sequence MFE | -31.76 |
| Consensus MFE | -25.76 |
| Energy contribution | -25.96 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5379799 112 - 22224390 AGAUAAAUCAAAAACCAUUUCCACCAACCGGACACACAGCGGACAAGGGGGCGUGGCCCCUUCGGUUGCGCCCCAUAAAAA-UGUCCUUCGAACUAUU-------CACCCCUUCGUGGCC ....................((((.((..((.........(((((..((((((..(((.....)))..)))))).......-)))))...((.....)-------).))..)).)))).. ( -33.10) >DroSec_CAF1 61137 107 - 1 ---UAAAUCAAAAGCCAUUUCCACCAACCGGACA--AAGCGGACAAGGGGGCGUGGCCCCUUCGGUUGCGCCCCAUAAAAA-UGUCCUUCGAACUAUU-------UACCCCUUCCUGGCC ---..........((((..(((.......)))..--....(((((..((((((..(((.....)))..)))))).......-)))))...........-------..........)))). ( -34.60) >DroSim_CAF1 48098 106 - 1 ---UAAAUCAAAUGCCAUUUCCACCAACCGGACA--AAGCGGACAAGG-GGCGUGGCCCCUUCGGUUGCGCCCCAUAAAAA-UGUCCUUCGAACUAUU-------CACCCCUUCCUGGCC ---..........((((..(((.......)))..--....(((((.((-((((..(((.....)))..)))))).......-)))))...........-------..........)))). ( -32.90) >DroEre_CAF1 54281 99 - 1 ---UAAAUCAAAAGCCAUGUCCAGCAGCCGGACA-----------AAGGGGCGUGGCCCCUUCGGUUGCGCCCCAUAAAAA-UGUCCUGCGAACUACUCACCAUUCACCCCUUC------ ---....((....((........)).((.(((((-----------..((((((..(((.....)))..)))))).......-))))).))))......................------ ( -31.10) >DroYak_CAF1 53732 102 - 1 ---UAAAUCAAAAGCCGUUUCCACCAACCGGACA-----------AGGGGGCGUGGCCCCUUCGGUUGCGCCCCAUAAAAACUGUCCUUCGAACUACU----ACUCACCUCUUCCUAGAC ---..........................(((((-----------..((((((..(((.....)))..))))))........)))))...........----.................. ( -27.10) >consensus ___UAAAUCAAAAGCCAUUUCCACCAACCGGACA___AGCGGACAAGGGGGCGUGGCCCCUUCGGUUGCGCCCCAUAAAAA_UGUCCUUCGAACUAUU_______CACCCCUUCCUGGCC .............................(((((.............((((((..(((.....)))..))))))........)))))................................. (-25.76 = -25.96 + 0.20)


| Location | 5,379,831 – 5,379,946 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.80 |
| Mean single sequence MFE | -38.86 |
| Consensus MFE | -30.96 |
| Energy contribution | -31.28 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5379831 115 - 22224390 UCAAAUUCGGUUGGGCAGCCAG-----CAGAAUGUUCGAUAGAUAAAUCAAAAACCAUUUCCACCAACCGGACACACAGCGGACAAGGGGGCGUGGCCCCUUCGGUUGCGCCCCAUAAAA .....(((((((((((.....)-----).........(((......)))..............)))))))))...............((((((..(((.....)))..))))))...... ( -38.50) >DroSec_CAF1 61169 109 - 1 UCAAAUUCGGUUGGGCAGCCAG-----CAGAAUGUUCGA----UAAAUCAAAAGCCAUUUCCACCAACCGGACA--AAGCGGACAAGGGGGCGUGGCCCCUUCGGUUGCGCCCCAUAAAA ....((((.(((((....))))-----).))))((((((----....))....((....(((.......)))..--..))))))...((((((..(((.....)))..))))))...... ( -38.00) >DroSim_CAF1 48130 108 - 1 UCAAAUUCGGCUGGGCAGCCAG-----CAGAAUGUUCGA----UAAAUCAAAUGCCAUUUCCACCAACCGGACA--AAGCGGACAAGG-GGCGUGGCCCCUUCGGUUGCGCCCCAUAAAA ....((((.(((((....))))-----).))))((((((----....))....((....(((.......)))..--..))))))..((-((((..(((.....)))..))))))...... ( -40.40) >DroEre_CAF1 54314 103 - 1 UCAAUUUCGGUUGGGCGGCCAGC--AGCAGAAUGUUCGA----UAAAUCAAAAGCCAUGUCCAGCAGCCGGACA-----------AAGGGGCGUGGCCCCUUCGGUUGCGCCCCAUAAAA .....((((((((((((((.(((--(......)))).((----....))....)))..)))))...))))))..-----------..((((((..(((.....)))..))))))...... ( -39.20) >DroYak_CAF1 53768 105 - 1 UCAAUUUCGGUUGGGCAGCCAGUGGAGUAAAAUGUUCGA----UAAAUCAAAAGCCGUUUCCACCAACCGGACA-----------AGGGGGCGUGGCCCCUUCGGUUGCGCCCCAUAAAA .....((((((((((...)).((((((.......((.((----....)).))......))))))))))))))..-----------..((((((..(((.....)))..))))))...... ( -38.22) >consensus UCAAAUUCGGUUGGGCAGCCAG_____CAGAAUGUUCGA____UAAAUCAAAAGCCAUUUCCACCAACCGGACA___AGCGGACAAGGGGGCGUGGCCCCUUCGGUUGCGCCCCAUAAAA .....((((((((((((...............)))..((........))..............)))))))))...............((((((..(((.....)))..))))))...... (-30.96 = -31.28 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:57 2006