
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,377,872 – 5,378,104 |
| Length | 232 |
| Max. P | 0.928068 |

| Location | 5,377,872 – 5,377,984 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.91 |
| Mean single sequence MFE | -38.81 |
| Consensus MFE | -27.87 |
| Energy contribution | -28.68 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5377872 112 - 22224390 UU--U--UU----GUUUUCGAGUGGAAUACCACGCCCAGUUGCCCACUUAACCCGCGAUUUGCUCAAUCCGUGAGGUUAAGGUUGGGAUUUGGGACUUGGAACUUGGAACUUGGCUGCGC ..--.--..----((..((((((......(((.((((((...(((((((((((((((..((....))..)))).)))))))..))))..))))).).)))........))))))..)).. ( -37.34) >DroSec_CAF1 59313 107 - 1 -------------UUUUUGGGGUGGAAUACCACGCCCAGUUGCCCACUUUACCCGCGAUUUGCUCAAUCCGUGAGGUUAAGGUUGGGAUUUGCGACUUGGAACUUGGAACUUGGCUGGGC -------------...(((((((((....)))).)))))..((((((((.(((((((..((....))..)))).))).)))((..((..((.(((........))).))))..))))))) ( -34.50) >DroSim_CAF1 46262 107 - 1 -------------UUUUUCGGGUGGAAUACCACGCCCAGUUGCCCACUUAACCCGCGAUUUGCUCAAUCCGUGAGGUUAAGGUUGGGAUUUGGGACUUGGAACUUGGAACUUGGCUGGGC -------------......(((((........)))))....((((((((((((((((..((....))..)))).)))))))((..((.((..((........))..)).))..))))))) ( -40.40) >DroEre_CAF1 52579 104 - 1 GUGCU--UUGGUUUCUGCUGGGCGAACUGCCACGCCCAGCUGCCCACUUAACCCGCGAUUUGCUCAAUCUGUGAGGUUAAGGUUGGGAUUUGGAAC--------------UUGGCUGGGC ..(((--..((((((.((((((((........))))))))..((((((((((((((((((.....)))).))).)))))))..))))....)))))--------------).)))..... ( -43.60) >DroYak_CAF1 51748 120 - 1 UUCUUGGUGGUUUUCUUUUGGGUGAAAUGCCACGCCCACUUGCCCACUUAACCCGCGAUUUGCACAAUCCGUGAGGUUAAGGUUGGGAUUUGGAACCCGGAACUUGGAAGUUGGCUGGUC ......(((((((((........)))).)))))(((.((((.((..(((((((((((..((....))..)))).))))))).(((((........))))).....)))))).)))..... ( -38.20) >consensus _U__U__U_____UUUUUCGGGUGGAAUACCACGCCCAGUUGCCCACUUAACCCGCGAUUUGCUCAAUCCGUGAGGUUAAGGUUGGGAUUUGGGACUUGGAACUUGGAACUUGGCUGGGC .....................(((......)))((((((((.(((((((((((((((..((....))..)))).)))))))..))))........((........)).....)))))))) (-27.87 = -28.68 + 0.81)



| Location | 5,377,912 – 5,378,024 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.46 |
| Mean single sequence MFE | -21.16 |
| Consensus MFE | -16.58 |
| Energy contribution | -16.66 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5377912 112 - 22224390 CUUGUUUUUCAGUUUUUUCUCCCUUUUUGUAUAUUUUUUUUU--U--UU----GUUUUCGAGUGGAAUACCACGCCCAGUUGCCCACUUAACCCGCGAUUUGCUCAAUCCGUGAGGUUAA ..........................................--.--..----......((((((...((........))...))))))((((((((..((....))..)))).)))).. ( -15.50) >DroSec_CAF1 59353 98 - 1 CUUGUUUUUCAGUUUUCACUUUCUUUUAUUA----------------------UUUUUGGGGUGGAAUACCACGCCCAGUUGCCCACUUUACCCGCGAUUUGCUCAAUCCGUGAGGUUAA ...............................----------------------...(((((((((....)))).)))))...........(((((((..((....))..)))).)))... ( -19.10) >DroSim_CAF1 46302 98 - 1 CUUGUUUUUCAGUUUUUUCUCGGUUUUUUUA----------------------UUUUUCGGGUGGAAUACCACGCCCAGUUGCCCACUUAACCCGCGAUUUGCUCAAUCCGUGAGGUUAA ...............................----------------------......(((((........)))))...........(((((((((..((....))..)))).))))). ( -19.10) >DroEre_CAF1 52605 110 - 1 CUUGUUUUUCAUGUUUUU---UAUUU-----AUUUUUCUUGUGCU--UUGGUUUCUGCUGGGCGAACUGCCACGCCCAGCUGCCCACUUAACCCGCGAUUUGCUCAAUCUGUGAGGUUAA ..................---.....-----.........(((..--..(((....((((((((........)))))))).)))))).((((((((((((.....)))).))).))))). ( -29.70) >DroYak_CAF1 51788 117 - 1 CUUGUUUUUCCAUUUUUC---CAUUUUUCCAAUUUUUCCUUUCUUGGUGGUUUUCUUUUGGGUGAAAUGCCACGCCCACUUGCCCACUUAACCCGCGAUUUGCACAAUCCGUGAGGUUAA ..................---........................(((((........((((((........)))))).....)))))(((((((((..((....))..)))).))))). ( -22.42) >consensus CUUGUUUUUCAGUUUUUUCU_CAUUUUUUUA__UUUU__U_U__U__U_____UUUUUCGGGUGGAAUACCACGCCCAGUUGCCCACUUAACCCGCGAUUUGCUCAAUCCGUGAGGUUAA ...........................................................(((((........)))))...........(((((((((..((....))..)))).))))). (-16.58 = -16.66 + 0.08)

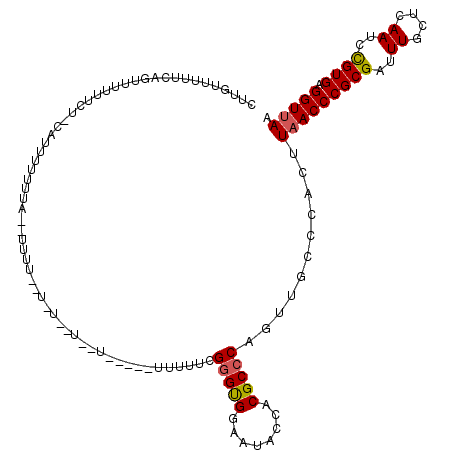
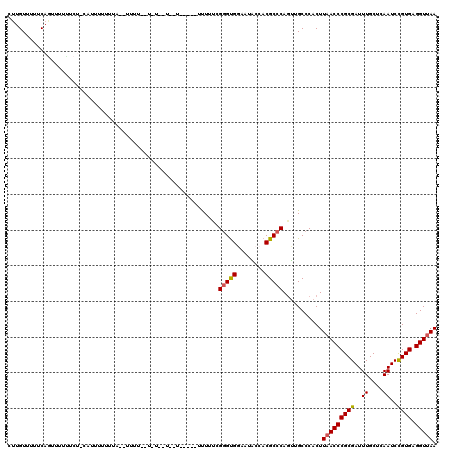
| Location | 5,377,952 – 5,378,064 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.59 |
| Mean single sequence MFE | -22.60 |
| Consensus MFE | -15.32 |
| Energy contribution | -14.76 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5377952 112 + 22224390 ACUGGGCGUGGUAUUCCACUCGAAAAC----AA--A--AAAAAAAAAUAUACAAAAAGGGAGAAAAAACUGAAAAACAAGUUCGGCGCGAAACGAAGAUAAAGUGGUUUGGCUUGGGGCU .(..(((((((....)))).((((.((----..--.--.............................(((........)))(((........))).......))..)))))))..).... ( -17.60) >DroSec_CAF1 59393 98 + 1 ACUGGGCGUGGUAUUCCACCCCAAAAA----------------------UAAUAAAAGAAAGUGAAAACUGAAAAACAAGUUCGGCGCGAAACGAAGAUAAAGUGGUUUGGCUUGGGGCU ..((((.((((....))))))))....----------------------...........(((....)))........(((((.(.((.((((.(........).)))).)).).))))) ( -24.60) >DroSim_CAF1 46342 98 + 1 ACUGGGCGUGGUAUUCCACCCGAAAAA----------------------UAAAAAAACCGAGAAAAAACUGAAAAACAAGUUCGGCGCGAAACGAAGAUAAAGUGGUUUGGCUUGGGGCU ....((.((((....))))))......----------------------........(((((...((((((.........((((........)))).......))))))..))))).... ( -21.09) >DroEre_CAF1 52645 110 + 1 GCUGGGCGUGGCAGUUCGCCCAGCAGAAACCAA--AGCACAAGAAAAAU-----AAAUA---AAAAACAUGAAAAACAAGUUCGGCGCGGAACGAAGACAAAGUGGUUUGGCUUGGGGCU ((((((((........)))))))).....((((--(.(((.........-----.....---.......((.....)).((((......)))).........))).)))))......... ( -30.10) >DroYak_CAF1 51828 117 + 1 AGUGGGCGUGGCAUUUCACCCAAAAGAAAACCACCAAGAAAGGAAAAAUUGGAAAAAUG---GAAAAAUGGAAAAACAAGUUCGCCGUGGAACGAAGAUAAAGUGAUUUGGCUUGGGGCU ....(((((((..((((........)))).)))).........................---(((...((......))..))))))..((..(.(((.((((....)))).))).)..)) ( -19.60) >consensus ACUGGGCGUGGUAUUCCACCCAAAAAA_____A__A__A_A__AAAA__UAAAAAAAGA_AGAAAAAACUGAAAAACAAGUUCGGCGCGAAACGAAGAUAAAGUGGUUUGGCUUGGGGCU ....((.((((....)))))).........................................................(((((.(.((.((((.(........).)))).)).).))))) (-15.32 = -14.76 + -0.56)

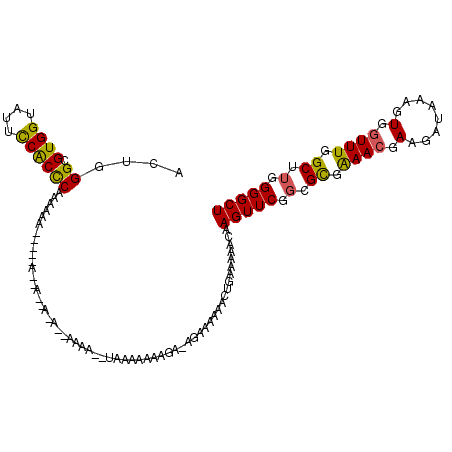

| Location | 5,377,984 – 5,378,104 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.74 |
| Mean single sequence MFE | -28.88 |
| Consensus MFE | -23.88 |
| Energy contribution | -23.72 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5377984 120 + 22224390 AAAAAAAUAUACAAAAAGGGAGAAAAAACUGAAAAACAAGUUCGGCGCGAAACGAAGAUAAAGUGGUUUGGCUUGGGGCUCUGUCACCGGAGCUUGGUUGCCUACCAGGAGCCCCAUAAA .................(((....................((((........)))).......((((..(((.(.((((((((....)))))))).)..))).)))).....)))..... ( -28.90) >DroSec_CAF1 59420 111 + 1 ---------UAAUAAAAGAAAGUGAAAACUGAAAAACAAGUUCGGCGCGAAACGAAGAUAAAGUGGUUUGGCUUGGGGCUCUGUGACCAAGUCUUGGUUGCCUACCAGGAGCCCCAUAAA ---------...........(((....)))................((.((((.(........).)))).)).((((((((((..((((.....))))..)......))))))))).... ( -31.30) >DroSim_CAF1 46369 111 + 1 ---------UAAAAAAACCGAGAAAAAACUGAAAAACAAGUUCGGCGCGAAACGAAGAUAAAGUGGUUUGGCUUGGGGCUCUGUCACCAAGUCUUGGUUGCCUACCAGGAGCCCCAUAAA ---------........((((......(((........))))))).((.((((.(........).)))).)).(((((((((((.((((.....)))).))......))))))))).... ( -29.80) >DroEre_CAF1 52683 112 + 1 AAGAAAAAU-----AAAUA---AAAAACAUGAAAAACAAGUUCGGCGCGGAACGAAGACAAAGUGGUUUGGCUUGGGGCUCUGUCACCAAGUCUUGGUUGCCUACCAGGAGCCCCAUAAA .........-----.....---........................((.((((.(........).)))).)).(((((((((((.((((.....)))).))......))))))))).... ( -26.50) >DroYak_CAF1 51868 117 + 1 AGGAAAAAUUGGAAAAAUG---GAAAAAUGGAAAAACAAGUUCGCCGUGGAACGAAGAUAAAGUGAUUUGGCUUGGGGCUCUGUCACCAAGUCCUGCUUGCCUACCAGGAGCCCCAUAAA ...................---(((...((......))..)))((((....((.........))....)))).(((((((((.....((((.....)))).......))))))))).... ( -27.90) >consensus A__AAAA__UAAAAAAAGA_AGAAAAAACUGAAAAACAAGUUCGGCGCGAAACGAAGAUAAAGUGGUUUGGCUUGGGGCUCUGUCACCAAGUCUUGGUUGCCUACCAGGAGCCCCAUAAA ..............................................((.((((.(........).)))).)).(((((((((((.((((.....)))).))......))))))))).... (-23.88 = -23.72 + -0.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:52 2006