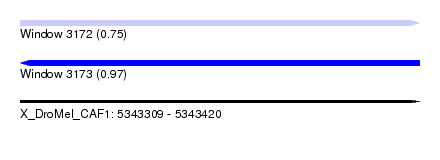
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,343,309 – 5,343,420 |
| Length | 111 |
| Max. P | 0.970505 |

| Location | 5,343,309 – 5,343,420 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 66.06 |
| Mean single sequence MFE | -23.17 |
| Consensus MFE | -11.48 |
| Energy contribution | -12.35 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5343309 111 + 22224390 UCUAAGAAAACGAAAUAUUCUUGCUUGUAACGAAUUACUAUAUAUGUAUGUACAUACAUUGCAU-UCCUUGCAAUCCACGAAGUGCUUGCAAAUGCAAGGAUAGCAGUUGAU-- ........(((....(((((((((..((((....))))......((((.((((....((((((.-....)))))).......)))).))))...)))))))))...)))...-- ( -23.90) >DroSec_CAF1 18811 96 + 1 ---------ACUAAAUGUUCUUGCUUGUAACGAAUUAUUAUAUAUGUGGG--------UUGCAU-UCCUUGCAACCAACGAUGGGUUUGUAAAUGCAAGGAUAGCAGUUGAUAA ---------(((...(((((((((...(.(((((((....(((.(((.((--------(((((.-....))))))).))))))))))))).)..)))))))))..)))...... ( -26.80) >DroSim_CAF1 16262 87 + 1 UCUAAGAAAACUAAAUGUUCUUGCUUGUAAGGAAAUAUUAUACAUGUGGG--------UUGCAU-UCCCUGCA------------------AAUGCAAGGAUAGCAGUUGAUAA ........((((...((((((((((((((.(((..........((((...--------..))))-))).))))------------------)..)))))))))..))))..... ( -19.50) >DroYak_CAF1 19375 102 + 1 UUCUAGAAAACGAAAUAUUCUUGCUACUGGCGAGUUACUAUACUA----G--------UUGCAUAUACUAGCAGCCCACGAAUUUUGUGCAAGUGCAAGGGGAGCAGUUUAGAA .((((((..........(((((((.(((.((((((......))).----(--------((((........)))))............))).))))))))))......)))))). ( -22.49) >consensus UCUAAGAAAACGAAAUAUUCUUGCUUGUAACGAAUUACUAUACAUGUGGG________UUGCAU_UCCUUGCAACCCACGAAGUGUUUGCAAAUGCAAGGAUAGCAGUUGAUAA ........(((....(((((((((.(((((.......)))))................(((((......)))))....................)))))))))...)))..... (-11.48 = -12.35 + 0.87)



| Location | 5,343,309 – 5,343,420 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 66.06 |
| Mean single sequence MFE | -20.11 |
| Consensus MFE | -11.98 |
| Energy contribution | -13.10 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
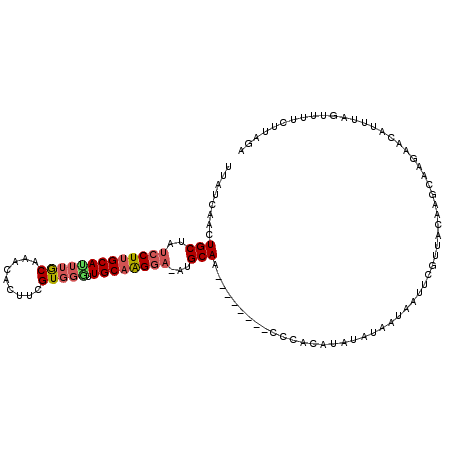
>X_DroMel_CAF1 5343309 111 - 22224390 --AUCAACUGCUAUCCUUGCAUUUGCAAGCACUUCGUGGAUUGCAAGGA-AUGCAAUGUAUGUACAUACAUAUAUAGUAAUUCGUUACAAGCAAGAAUAUUUCGUUUUCUUAGA --.......(((.((((((((((..(.........)..)).))))))))-.(((.((((((((....)))))))).)))..........)))(((((.........)))))... ( -27.20) >DroSec_CAF1 18811 96 - 1 UUAUCAACUGCUAUCCUUGCAUUUACAAACCCAUCGUUGGUUGCAAGGA-AUGCAA--------CCCACAUAUAUAAUAAUUCGUUACAAGCAAGAACAUUUAGU--------- ...............(((((...............((.(((((((....-.)))))--------)).)).....((((.....))))...)))))..........--------- ( -17.60) >DroSim_CAF1 16262 87 - 1 UUAUCAACUGCUAUCCUUGCAUU------------------UGCAGGGA-AUGCAA--------CCCACAUGUAUAAUAUUUCCUUACAAGCAAGAACAUUUAGUUUUCUUAGA ........((((.(((((((...------------------.)))))))-(((((.--------......)))))..............))))(((((.....)))))...... ( -17.00) >DroYak_CAF1 19375 102 - 1 UUCUAAACUGCUCCCCUUGCACUUGCACAAAAUUCGUGGGCUGCUAGUAUAUGCAA--------C----UAGUAUAGUAACUCGCCAGUAGCAAGAAUAUUUCGUUUUCUAGAA .(((((((.......((((((((..(((.......)))((((((((((.......)--------)----)))))..(.....))))))).)))))........)))...)))). ( -18.66) >consensus UUAUCAACUGCUAUCCUUGCAUUUGCAAACACUUCGUGGGUUGCAAGGA_AUGCAA________CCCACAUAUAUAAUAAUUCGUUACAAGCAAGAACAUUUAGUUUUCUUAGA ........(((..(((((((((((((.........))))).))))))))...)))........................................................... (-11.98 = -13.10 + 1.12)

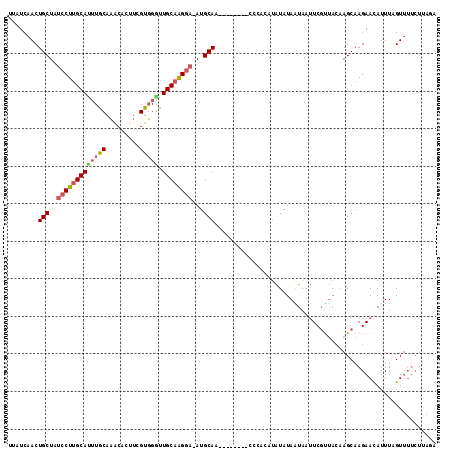
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:39 2006