
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,319,373 – 5,319,586 |
| Length | 213 |
| Max. P | 0.938296 |

| Location | 5,319,373 – 5,319,493 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.18 |
| Mean single sequence MFE | -36.83 |
| Consensus MFE | -31.29 |
| Energy contribution | -31.30 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5319373 120 + 22224390 GGUGUUGGGGGGCUAGAGAAAAUCCACCUUGGCCAGAGGCCAUCCAGCUAUUAUUGCUGAUGCCUUUUUAUUUUAAUUCCCAAAUCGAAAAAGUUUCGCCUUGGCGAUUAGGCGAGUAAA (((.((((((((((((.(........).))))))((((((.(((.(((.......))))))))))))..........)))))))))........(((((((........))))))).... ( -37.20) >DroSim_CAF1 47284 118 + 1 GGGGU--GGGGGCCAGAGAAAAUCCUACUUGGCCAGAGGCCAUCCAGCUAUUAUUGCUGAUGCCUUUUUAUUUUAAUUCCCAAAUCGAAAAAGUUUCGCCCUGGCGAUUAGGCGAGUAAA ((..(--((((((((.((......))...)))))((((((.(((.(((.......))))))))))))...........))))..))........((((((..........)))))).... ( -36.60) >DroEre_CAF1 58460 118 + 1 GUGGA--GGGGGCUAGAGUAAAUCCUCCCUGGCCAGAGGCCAUUUAGCUAUUAUUGCUGAUGCCUUUUUAUUUUAAUUCCCAAAUCGAAAAAGUUUCGCCUUGGCGAUUAGGCAAGUAAA ..(((--(((.((....))...))))))((.(((((((((...(((((.......))))).))))))......(((((.((((..((((.....))))..)))).)))))))).)).... ( -36.70) >consensus GGGGU__GGGGGCUAGAGAAAAUCCUCCUUGGCCAGAGGCCAUCCAGCUAUUAUUGCUGAUGCCUUUUUAUUUUAAUUCCCAAAUCGAAAAAGUUUCGCCUUGGCGAUUAGGCGAGUAAA .......(((((((((............))))))((((((.(((.(((.......))))))))))))...........))).............(((((((........))))))).... (-31.29 = -31.30 + 0.01)

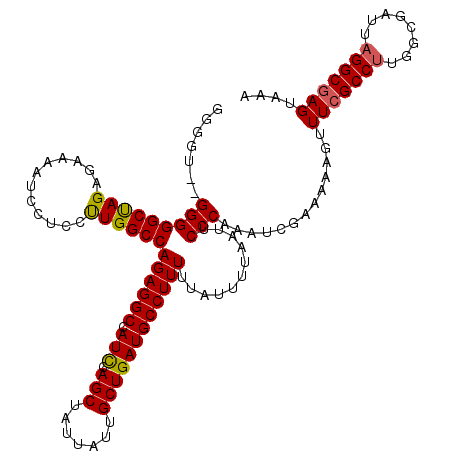
| Location | 5,319,413 – 5,319,533 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -30.37 |
| Consensus MFE | -30.58 |
| Energy contribution | -30.03 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.92 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5319413 120 - 22224390 UUUCCGCAGCUUUUCCAAGAAACGCUUAAAGCGUUUAGAGUUUACUCGCCUAAUCGCCAAGGCGAAACUUUUUCGAUUUGGGAAUUAAAAUAAAAAGGCAUCAGCAAUAAUAGCUGGAUG ......(((((((((((((((((((.....))))))((((((...((((((........)))))))))))).....)))))))).............((....))......))))).... ( -32.00) >DroSim_CAF1 47322 118 - 1 --UGCACAGCUUUUCCAAGAAACGCUUAAAGCGUUUAGAGUUUACUCGCCUAAUCGCCAGGGCGAAACUUUUUCGAUUUGGGAAUUAAAAUAAAAAGGCAUCAGCAAUAAUAGCUGGAUG --....(((((((((((((((((((.....))))))((((((...((((((........)))))))))))).....)))))))).............((....))......))))).... ( -32.30) >DroEre_CAF1 58498 120 - 1 UUUGCACGGCUUUUCCAAGAAGCGCUUAAAGCGUUUAGAGUUUACUUGCCUAAUCGCCAAGGCGAAACUUUUUCGAUUUGGGAAUUAAAAUAAAAAGGCAUCAGCAAUAAUAGCUAAAUG .......((((((((((((((((((.....))))))((((((...((((((........)))))))))))).....)))))))).............((....))......))))..... ( -26.80) >consensus UUUGCACAGCUUUUCCAAGAAACGCUUAAAGCGUUUAGAGUUUACUCGCCUAAUCGCCAAGGCGAAACUUUUUCGAUUUGGGAAUUAAAAUAAAAAGGCAUCAGCAAUAAUAGCUGGAUG ......(((((((((((((((((((.....))))))((((((...((((((........)))))))))))).....)))))))).............((....))......))))).... (-30.58 = -30.03 + -0.55)



| Location | 5,319,453 – 5,319,546 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.77 |
| Mean single sequence MFE | -21.10 |
| Consensus MFE | -17.66 |
| Energy contribution | -17.00 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.58 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.606295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5319453 93 - 22224390 UUAGCCC---------------------------ACCGAUUUUCCGCAGCUUUUCCAAGAAACGCUUAAAGCGUUUAGAGUUUACUCGCCUAAUCGCCAAGGCGAAACUUUUUCGAUUUG ...(((.---------------------------..(((((....((............((((((.....)))))).(((....)))))..)))))....)))................. ( -19.70) >DroSim_CAF1 47362 80 - 1 UU----------------------------------------UGCACAGCUUUUCCAAGAAACGCUUAAAGCGUUUAGAGUUUACUCGCCUAAUCGCCAGGGCGAAACUUUUUCGAUUUG ..----------------------------------------.................((((((.....))))))((((((...((((((........))))))))))))......... ( -17.70) >DroEre_CAF1 58538 120 - 1 UUUGCCCAGUCCCGAAUUCGUCGCCAAAUUUGCCAACGAUUUUGCACGGCUUUUCCAAGAAGCGCUUAAAGCGUUUAGAGUUUACUUGCCUAAUCGCCAAGGCGAAACUUUUUCGAUUUG ............((((....(((((....(((....(((((..(((.(....(((....((((((.....)))))).)))....).)))..))))).))))))))......))))..... ( -25.90) >consensus UU_GCCC___________________________A_CGAUUUUGCACAGCUUUUCCAAGAAACGCUUAAAGCGUUUAGAGUUUACUCGCCUAAUCGCCAAGGCGAAACUUUUUCGAUUUG ...........................................................((((((.....))))))((((((...((((((........))))))))))))......... (-17.66 = -17.00 + -0.66)



| Location | 5,319,493 – 5,319,586 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.77 |
| Mean single sequence MFE | -23.80 |
| Consensus MFE | -15.55 |
| Energy contribution | -15.33 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5319493 93 + 22224390 CUCUAAACGCUUUAAGCGUUUCUUGGAAAAGCUGCGGAAAAUCGGU---------------------------GGGCUAAAGCGUAUGUGAAAUACUAAUUUAACAUGUCUGAGAAGUUU (((...((((((((.((....(((....)))(..(.(.....).).---------------------------.)))))))))))((((.((((....)))).))))....)))...... ( -21.20) >DroSim_CAF1 47402 80 + 1 CUCUAAACGCUUUAAGCGUUUCUUGGAAAAGCUGUGCA----------------------------------------AAAGCGUAUGUGAAAUACUAAUUUAACAUGUCUGAGAACUUU .....(((((.....)))))(((..((...(((.....----------------------------------------..))).(((((.((((....)))).)))))))..)))..... ( -17.40) >DroEre_CAF1 58578 120 + 1 CUCUAAACGCUUUAAGCGCUUCUUGGAAAAGCCGUGCAAAAUCGUUGGCAAAUUUGGCGACGAAUUCGGGACUGGGCAAAGGCGUAUGUGAAAUACUAAUUUAACAUGUCUGAGAAGUUU ........((.....))((((((..((...(((.(((....((((((.((....)).))))))..(((....))))))..))).(((((.((((....)))).)))))))..)))))).. ( -32.80) >consensus CUCUAAACGCUUUAAGCGUUUCUUGGAAAAGCUGUGCAAAAUCG_U___________________________GGGC_AAAGCGUAUGUGAAAUACUAAUUUAACAUGUCUGAGAAGUUU .......(((.....)))(((((..((...(((...............................................))).(((((.((((....)))).)))))))..)))))... (-15.55 = -15.33 + -0.22)

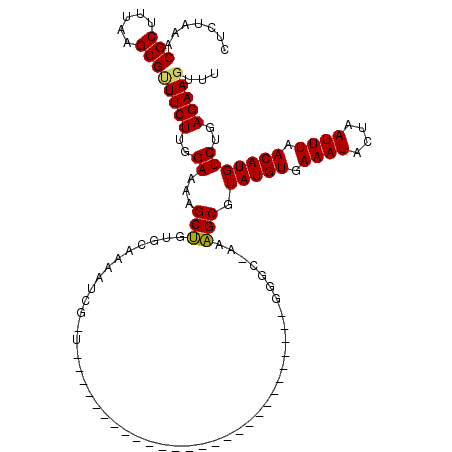

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:35 2006