
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,318,451 – 5,318,651 |
| Length | 200 |
| Max. P | 0.869701 |

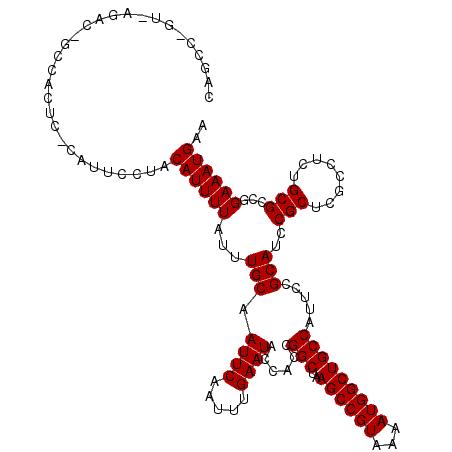
| Location | 5,318,451 – 5,318,571 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.38 |
| Mean single sequence MFE | -28.16 |
| Consensus MFE | -21.64 |
| Energy contribution | -21.64 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5318451 120 - 22224390 CAGCCGGUGAGACCGCCACUCUCAUUCCUACAUUUUAUUUGCAAUUCAAUUUGAAUAACACCGGCUAAGCCGUAAAAUGGCUGCCAUUCCGCAUCCGCUCGCCUCUGCGCCGGAAAUGAA .(((((((((((.......))))....................((((.....))))...))))))).((((((...))))))..(((((((....(((........))).))).)))).. ( -31.70) >DroSec_CAF1 54674 107 - 1 CAGC------------CACUC-CAUUUCUCCAUUUUAUUUGCAAUUCAAUUUGAAUACCACCGGCUAAGCCGUAAAAUGGCUGCCAUUCCGCAUCCGCUCGCCUCUGCGCCGGAAAUGAA ....------------.....-(((((((..........(((.((((.....))))......(((..((((((...))))))))).....)))..(((........)))..))))))).. ( -24.20) >DroSim_CAF1 46154 107 - 1 CAGC------------CACUC-CAUUUCUCCAUUUUAUUUGCAAUUCAAUUUGAAUACCACCGGCUAAGCCGUAAAAUGGCUGCCAUUCCGCAUCCGCUCGCCUCUGCGCCGGAAAUGAA ....------------.....-(((((((..........(((.((((.....))))......(((..((((((...))))))))).....)))..(((........)))..))))))).. ( -24.20) >DroEre_CAF1 57570 119 - 1 CAGCCGGUAAGACUGCGGCUC-CAUUCCAGCAUUUUAUUUGCAAUUCAAUUUGAAUACCAACGGCUAAGCCGUAAAAUGGCUGCCAUUCCGCAUCCGCUUUCCUCCGCGCCGGAAAUGAA ((.(((((..((.(((((...-.......(((.......))).((((.....))))......(((..((((((...)))))))))...))))))).((........)))))))...)).. ( -32.50) >DroYak_CAF1 57988 115 - 1 ----CAGUGAGACUGCCACUC-CAUGCCCACAUUUUAUUUGCAAUUCAAUUUGAAUACCGCCGGCUAAGCCGUAAAAUGGCUGCCAUUCCGCAUCCGCUUGGCUCUGCGCCGGAAAUGAA ----.((((.......)))).-........((((((...((((((((.....))))...((((((..((((((...))))))))).....((....))..)))..))))...)))))).. ( -28.20) >consensus CAGCC_GU_AGAC_GCCACUC_CAUUCCUACAUUUUAUUUGCAAUUCAAUUUGAAUACCACCGGCUAAGCCGUAAAAUGGCUGCCAUUCCGCAUCCGCUCGCCUCUGCGCCGGAAAUGAA ..............................((((((...(((.((((.....))))......(((..((((((...))))))))).....)))..(((........)))...)))))).. (-21.64 = -21.64 + 0.00)


| Location | 5,318,491 – 5,318,611 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.27 |
| Mean single sequence MFE | -28.89 |
| Consensus MFE | -15.08 |
| Energy contribution | -16.08 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.720522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5318491 120 + 22224390 CCAUUUUACGGCUUAGCCGGUGUUAUUCAAAUUGAAUUGCAAAUAAAAUGUAGGAAUGAGAGUGGCGGUCUCACCGGCUGCAGCAGCUGCUUAAAUAAUACAAUUCUAUUUGAAUGAUUU ........((((...))))..(((((((((((.((((((..........((((..((....)).((((((.....)))))).....))))..........)))))).))))))))))).. ( -32.35) >DroSec_CAF1 54714 107 + 1 CCAUUUUACGGCUUAGCCGGUGGUAUUCAAAUUGAAUUGCAAAUAAAAUGGAGAAAUG-GAGUG------------GCUGCAGCAGCUGCUUAAAUAAUACAAUUCUAUUUGAAUGAUUC ((((....((((...))))))))(((((((((.((((((............(....).-(((..------------((((...))))..)))........)))))).))))))))).... ( -28.60) >DroSim_CAF1 46194 107 + 1 CCAUUUUACGGCUUAGCCGGUGGUAUUCAAAUUGAAUUGCAAAUAAAAUGGAGAAAUG-GAGUG------------GCUGCAGCAGCUGCUUAAAUAAUACAAUUCUAUUUGAAUGAUUC ((((....((((...))))))))(((((((((.((((((............(....).-(((..------------((((...))))..)))........)))))).))))))))).... ( -28.60) >DroEre_CAF1 57610 119 + 1 CCAUUUUACGGCUUAGCCGUUGGUAUUCAAAUUGAAUUGCAAAUAAAAUGCUGGAAUG-GAGCCGCAGUCUUACCGGCUGCAGCAGCUGCUUAAAUAAUACAAUUCUAUUUGAAUGAUUU (((....(((((...))))))))(((((((((.((((((..........((((...((-.(((((.........))))).)).)))).............)))))).))))))))).... ( -33.90) >DroYak_CAF1 58028 91 + 1 CCAUUUUACGGCUUAGCCGGCGGUAUUCAAAUUGAAUUGCAAAUAAAAUGUGGGCAUG-GAGUGGCAGUCUCACUG----------------------------UCUAUUUGAAUGAUUC .(((((((((((...))))(((..((((.....)))))))...)))))))....(((.-(((((((((.....)))----------------------------.))))))..))).... ( -21.00) >consensus CCAUUUUACGGCUUAGCCGGUGGUAUUCAAAUUGAAUUGCAAAUAAAAUGGAGGAAUG_GAGUGGC_GUCU_AC_GGCUGCAGCAGCUGCUUAAAUAAUACAAUUCUAUUUGAAUGAUUC ........((((...))))....(((((((((.((((((........((......))...................((.((....)).))..........)))))).))))))))).... (-15.08 = -16.08 + 1.00)


| Location | 5,318,491 – 5,318,611 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.27 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -16.42 |
| Energy contribution | -17.96 |
| Covariance contribution | 1.54 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5318491 120 - 22224390 AAAUCAUUCAAAUAGAAUUGUAUUAUUUAAGCAGCUGCUGCAGCCGGUGAGACCGCCACUCUCAUUCCUACAUUUUAUUUGCAAUUCAAUUUGAAUAACACCGGCUAAGCCGUAAAAUGG .....((((((((.((((((((........((((...))))....((((....))))......................)))))))).)))))))).....((((...))))........ ( -28.60) >DroSec_CAF1 54714 107 - 1 GAAUCAUUCAAAUAGAAUUGUAUUAUUUAAGCAGCUGCUGCAGC------------CACUC-CAUUUCUCCAUUUUAUUUGCAAUUCAAUUUGAAUACCACCGGCUAAGCCGUAAAAUGG .....((((((((.((((((((........((((...)))).(.------------.....-)................)))))))).)))))))).(((.((((...)))).....))) ( -21.00) >DroSim_CAF1 46194 107 - 1 GAAUCAUUCAAAUAGAAUUGUAUUAUUUAAGCAGCUGCUGCAGC------------CACUC-CAUUUCUCCAUUUUAUUUGCAAUUCAAUUUGAAUACCACCGGCUAAGCCGUAAAAUGG .....((((((((.((((((((........((((...)))).(.------------.....-)................)))))))).)))))))).(((.((((...)))).....))) ( -21.00) >DroEre_CAF1 57610 119 - 1 AAAUCAUUCAAAUAGAAUUGUAUUAUUUAAGCAGCUGCUGCAGCCGGUAAGACUGCGGCUC-CAUUCCAGCAUUUUAUUUGCAAUUCAAUUUGAAUACCAACGGCUAAGCCGUAAAAUGG .....((((((((.((((((((.....((((..((((.((.(((((.........))))).-))...))))..))))..)))))))).)))))))).((((((((...)))))....))) ( -34.80) >DroYak_CAF1 58028 91 - 1 GAAUCAUUCAAAUAGA----------------------------CAGUGAGACUGCCACUC-CAUGCCCACAUUUUAUUUGCAAUUCAAUUUGAAUACCGCCGGCUAAGCCGUAAAAUGG .....((((((((.((----------------------------.((((.......)))).-..(((.............)))..)).)))))))).....((((...))))........ ( -13.12) >consensus GAAUCAUUCAAAUAGAAUUGUAUUAUUUAAGCAGCUGCUGCAGCC_GU_AGAC_GCCACUC_CAUUCCUACAUUUUAUUUGCAAUUCAAUUUGAAUACCACCGGCUAAGCCGUAAAAUGG .....((((((((.((((((((........((((...))))...(((.....)))........................)))))))).)))))))).....((((...))))........ (-16.42 = -17.96 + 1.54)



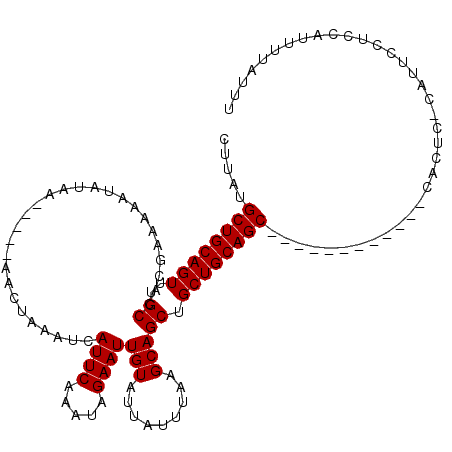
| Location | 5,318,531 – 5,318,651 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.62 |
| Mean single sequence MFE | -22.18 |
| Consensus MFE | -15.52 |
| Energy contribution | -15.53 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5318531 120 - 22224390 CUUAUGCUGCAGUUUGCAACGAAAAAAAAAAACAGAAACUAAAUCAUUCAAAUAGAAUUGUAUUAUUUAAGCAGCUGCUGCAGCCGGUGAGACCGCCACUCUCAUUCCUACAUUUUAUUU .....((((((((((((.................((.......))((((.....))))............))))..)))))))).((((....))))....................... ( -22.10) >DroSec_CAF1 54754 102 - 1 CUUAUGCUGCAGUUUGCCACGAAAAAUAUAA-----AACUGAAUCAUUCAAAUAGAAUUGUAUUAUUUAAGCAGCUGCUGCAGC------------CACUC-CAUUUCUCCAUUUUAUUU .....((((((((((((...(((.(((((((-----..(((...........)))..))))))).)))..))))..))))))))------------.....-.................. ( -19.10) >DroSim_CAF1 46234 102 - 1 CUUAUGCUGCAGUUUGCCACGAAAAAUAUAA-----AACUGAAUCAUUCAAAUAGAAUUGUAUUAUUUAAGCAGCUGCUGCAGC------------CACUC-CAUUUCUCCAUUUUAUUU .....((((((((((((...(((.(((((((-----..(((...........)))..))))))).)))..))))..))))))))------------.....-.................. ( -19.10) >DroEre_CAF1 57650 112 - 1 CUUAUGCUGCAGUUUGCCACGAACAA-AUA------AUCCAAAUCAUUCAAAUAGAAUUGUAUUAUUUAAGCAGCUGCUGCAGCCGGUAAGACUGCGGCUC-CAUUCCAGCAUUUUAUUU .....(((((((((((((......((-(((------((.(((.((.........)).))).)))))))..((((...))))....))).))))))))))..-.................. ( -28.40) >consensus CUUAUGCUGCAGUUUGCCACGAAAAAUAUAA_____AACUAAAUCAUUCAAAUAGAAUUGUAUUAUUUAAGCAGCUGCUGCAGC____________CACUC_CAUUCCUCCAUUUUAUUU .....((((((((..((............................((((.....))))(((.........))))).)))))))).................................... (-15.52 = -15.53 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:30 2006