
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,316,649 – 5,316,804 |
| Length | 155 |
| Max. P | 0.986027 |

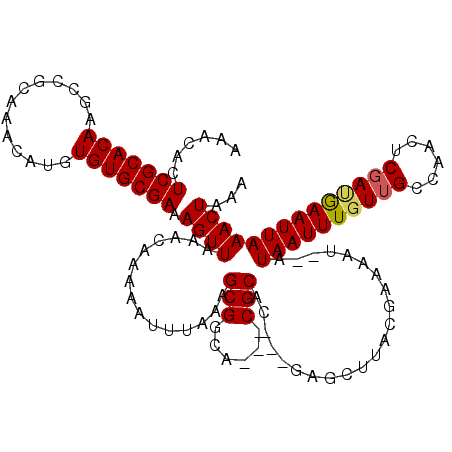
| Location | 5,316,649 – 5,316,767 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -22.35 |
| Consensus MFE | -18.54 |
| Energy contribution | -18.62 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5316649 118 - 22224390 AAACACUCGCACAAGCCGCAAACAUGUGUGCGAAAGUUAAACAAAAAUUUAAAGCGGCACCACGCACAGAAAGCUUACGAAAAU--AUAAUUUGUGGCCAACUCGAUGAAUUAAACUAAA ...((.(((.....((((((((..(((..((....))...)))..........(((......)))...................--....)))))))).....)))))............ ( -23.30) >DroSec_CAF1 53017 112 - 1 AAACACUCGCACAAACCGCAAACAUGUGUGCGAAAGUUAAACAAAAAUUUAAAGCGGCA---CGCAC---GAGCUUACGAAAAU--AUAAUUUGUUGCCAACUCGAUGAAUUAAACUAAA .....(((((.......)).....(((((((.....(((((......)))))....)))---)))).---)))...........--.(((((..(((......)))..)))))....... ( -19.10) >DroSim_CAF1 44493 112 - 1 AAACACUCGCACAAGCCGCAAACAUGUGUGCGAAAGUUAAACAAAAAUUUAAAGCGGCA---CGCAC---GAGCUUACGAAAAU--AUAAUUUGUUGCCAACUCGACGAAUUAAACUAAA .....((((.....(((((.....(((..((....))...)))..........))))).---....)---)))...........--.((((((((((......))))))))))....... ( -23.66) >DroEre_CAF1 56032 112 - 1 AAACACUCGCACAAGCCGCAAACAUGUGUGCGAAAGUUAAACAAAAAUUUAAAGCGGCA---CGCAC---GAGCUUACGAAAAU--AUAAUUUGUUGCCAACUCGACGAAUUAAACUGAA .....((((.....(((((.....(((..((....))...)))..........))))).---....)---)))...........--.((((((((((......))))))))))....... ( -23.66) >DroYak_CAF1 56199 114 - 1 AAACACUCGCACAAGCCGCAAACAUGUGUGCGAAAGUUAAACAAAAAUUUAAAGCGGCA---CGCAC---GAGCCUGUGAAAAUAUAUAAUUUUUUGCCAACUCGAUAAAUUAAACUAAA .(((..(((((((.............)))))))..))).......((((((..(((...---))).(---(((...(..((((........))))..)...)))).))))))........ ( -22.02) >consensus AAACACUCGCACAAGCCGCAAACAUGUGUGCGAAAGUUAAACAAAAAUUUAAAGCGGCA___CGCAC___GAGCUUACGAAAAU__AUAAUUUGUUGCCAACUCGAUGAAUUAAACUAAA ......(((((((.............))))))).((((...............(((......)))......................((((((((((......))))))))))))))... (-18.54 = -18.62 + 0.08)

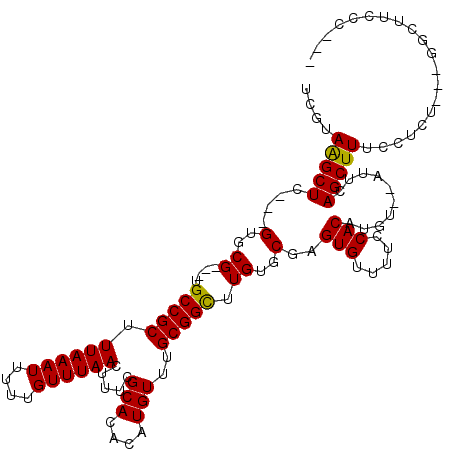
| Location | 5,316,687 – 5,316,804 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.70 |
| Mean single sequence MFE | -27.04 |
| Consensus MFE | -19.66 |
| Energy contribution | -19.34 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5316687 117 + 22224390 UCGUAAGCUUUCUGUGCGUGGUGCCGCUUUAAAUUUUUGUUUAACUUUCGCACACAUGUUUGCGGCUUGUGCGAGUGUUUUCCACAUGUAUAUUCAGCUUUCCUCU---GGCUUCCCCAU .(((((((....((((((((....))).((((((....)))))).....)))))...)))))))(..((((((.(((.....))).))))))..)((((.......---))))....... ( -28.50) >DroSec_CAF1 53055 106 + 1 UCGUAAGCUC---GUGCG---UGCCGCUUUAAAUUUUUGUUUAACUUUCGCACACAUGUUUGCGGUUUGUGCGAGUGUUUUCCACAUGU--AUUCAGCUUUCCUCU---GGCUUCCC--- ......((((---(..((---.(((((.((((((....)))))).....(((....)))..))))).))..))))).............--....((((.......---))))....--- ( -24.90) >DroSim_CAF1 44531 106 + 1 UCGUAAGCUC---GUGCG---UGCCGCUUUAAAUUUUUGUUUAACUUUCGCACACAUGUUUGCGGCUUGUGCGAGUGUUUUCCACAUGU--AUUCAGCUUUCCUCU---GGCUUCCC--- ......((((---(..((---.(((((.((((((....)))))).....(((....)))..))))).))..))))).............--....((((.......---))))....--- ( -26.40) >DroEre_CAF1 56070 109 + 1 UCGUAAGCUC---GUGCG---UGCCGCUUUAAAUUUUUGUUUAACUUUCGCACACAUGUUUGCGGCUUGUGCGAGUGUUUUCCACAUGU--AUUCAGCUUCCCGUUUUGCGUUUUCC--- .(((((((.(---(((.(---(((....((((((....)))))).....)))).))))...))((((.(((((.(((.....))).)))--))..)))).......)))))......--- ( -26.60) >DroYak_CAF1 56239 106 + 1 UCACAGGCUC---GUGCG---UGCCGCUUUAAAUUUUUGUUUAACUUUCGCACACAUGUUUGCGGCUUGUGCGAGUGUUUUUCACAUGU--AUUUAGCUUGCCUCU---GGUUUUCC--- (((.((((((---(..((---.(((((.((((((....)))))).....(((....)))..))))).))..)))(((.....)))....--.........)))).)---))......--- ( -28.80) >consensus UCGUAAGCUC___GUGCG___UGCCGCUUUAAAUUUUUGUUUAACUUUCGCACACAUGUUUGCGGCUUGUGCGAGUGUUUUCCACAUGU__AUUCAGCUUUCCUCU___GGCUUCCC___ ....(((((....(..((....(((((.((((((....)))))).....(((....)))..))))).))..)..(((.....)))..........))))).................... (-19.66 = -19.34 + -0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:26 2006