
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,316,027 – 5,316,132 |
| Length | 105 |
| Max. P | 0.948689 |

| Location | 5,316,027 – 5,316,132 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 68.36 |
| Mean single sequence MFE | -23.63 |
| Consensus MFE | -18.39 |
| Energy contribution | -18.20 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5316027 105 + 22224390 AUAUAUGUGUGUACAUUUUCUUU----UGAGCCCCUAGUGGCUGGCGACAAGAGCCAAGUAUUU----------UCGGAUUUUUAGCCAUUUCGGGCAUAUUUUUUCUCUGCUGCUUUC ..........(((((...((...----.))((((..((((((((((.......)))..(.....----------.)........)))))))..))))............)).))).... ( -22.00) >DroSec_CAF1 52462 80 + 1 ----------AUACACUUGCUUU----UGAGCCCCUAGUGGCUGGCGACAAGAGCCAAGUAU------------UCGGAUUUUCAGCCAACUCGGGCAUCAUUUUU------------- ----------....(((((((((----((.(((((....))..)))..)))))).)))))..------------...........(((......))).........------------- ( -21.40) >DroSim_CAF1 43945 80 + 1 ----------AUACACUUGCUUU----UGAGCCCCUAGUGGCUGGCGACAAGAGCCAAGUAU------------UCGGAUUUUCAGCCAACUCGGGCAUCAUUUUU------------- ----------....(((((((((----((.(((((....))..)))..)))))).)))))..------------...........(((......))).........------------- ( -21.40) >DroEre_CAF1 55410 108 + 1 AUACAU----AUAUGUAUUUUUUUGGUUGAGCCCCUAGUGGCUGGCGACAAAAGCCAAACAUUUUGUGAAACACUUGGAUUUGCAGCCAGCUCGGGCAUAUUUU----C-G--GCUAUU (((((.----...))))).....(((((((((((.....(((((((..((((..((((.......(((...))))))).))))..))))))).))))......)----)-)--)))).. ( -29.71) >consensus __________AUACACUUGCUUU____UGAGCCCCUAGUGGCUGGCGACAAGAGCCAAGUAU____________UCGGAUUUUCAGCCAACUCGGGCAUAAUUUUU_____________ ..............................((((....((((((((.......))).....................(.....))))))....))))...................... (-18.39 = -18.20 + -0.19)

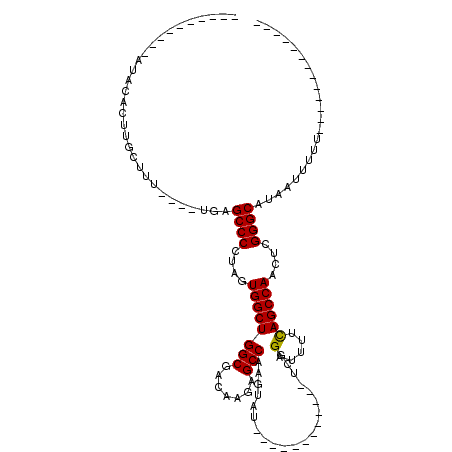

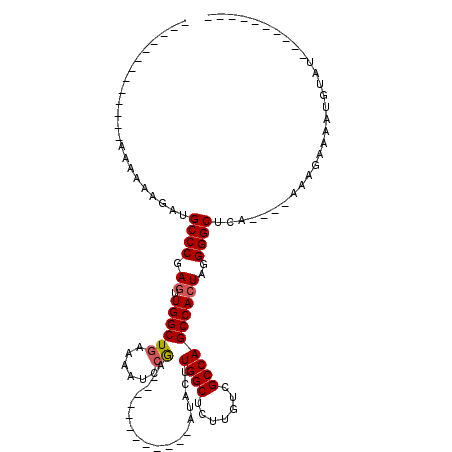
| Location | 5,316,027 – 5,316,132 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 68.36 |
| Mean single sequence MFE | -22.75 |
| Consensus MFE | -16.26 |
| Energy contribution | -16.57 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5316027 105 - 22224390 GAAAGCAGCAGAGAAAAAAUAUGCCCGAAAUGGCUAAAAAUCCGA----------AAAUACUUGGCUCUUGUCGCCAGCCACUAGGGGCUCA----AAAGAAAAUGUACACACAUAUAU ....((.((((((.........(((......))).......((((----------......))))))).))).)).((((......))))..----.......((((....)))).... ( -17.60) >DroSec_CAF1 52462 80 - 1 -------------AAAAAUGAUGCCCGAGUUGGCUGAAAAUCCGA------------AUACUUGGCUCUUGUCGCCAGCCACUAGGGGCUCA----AAAGCAAGUGUAU---------- -------------.....(((.((((.(((.(((((.......(.------------...)..(((....)))..))))))))..)))))))----.............---------- ( -21.20) >DroSim_CAF1 43945 80 - 1 -------------AAAAAUGAUGCCCGAGUUGGCUGAAAAUCCGA------------AUACUUGGCUCUUGUCGCCAGCCACUAGGGGCUCA----AAAGCAAGUGUAU---------- -------------.....(((.((((.(((.(((((.......(.------------...)..(((....)))..))))))))..)))))))----.............---------- ( -21.20) >DroEre_CAF1 55410 108 - 1 AAUAGC--C-G----AAAAUAUGCCCGAGCUGGCUGCAAAUCCAAGUGUUUCACAAAAUGUUUGGCUUUUGUCGCCAGCCACUAGGGGCUCAACCAAAAAAAUACAUAU----AUGUAU ......--.-.----.......((((..((((((.(((((.(((((..(((....)))..)))))..))))).))))))......))))............(((((...----.))))) ( -31.00) >consensus _____________AAAAAAGAUGCCCGAGUUGGCUGAAAAUCCGA____________AUACUUGGCUCUUGUCGCCAGCCACUAGGGGCUCA____AAAGAAAAUGUAU__________ ......................((((.((.((((((......))..................((((.......))))))))))..)))).............................. (-16.26 = -16.57 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:24 2006