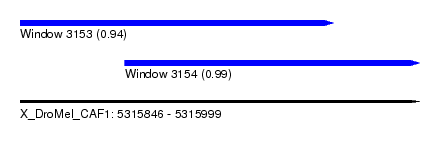
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,315,846 – 5,315,999 |
| Length | 153 |
| Max. P | 0.988847 |

| Location | 5,315,846 – 5,315,966 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -37.76 |
| Consensus MFE | -29.80 |
| Energy contribution | -29.60 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5315846 120 + 22224390 UCAAUUAGCGCUCAGAUCCUUGGACCAAUUCGCCUAUUUUGGCUGCUUAAUUUUGCGUAAAAUCGCACAGGUGACCGGCGAUACUGGCAACUAUUUCAAUUUGGCAGCCAAGUGGCUUUC ..............(.((....)).).....(((...((((((((((.((((.((((......))))..(((..((((.....))))..))).....)))).)))))))))).))).... ( -33.70) >DroSec_CAF1 52276 120 + 1 UCAAUUAGCGGUCAGAUCCUUGGGCCAAUUCGACUAUUUUGGCUGCUUAAUUUUGCGUAAAAUCGCACAGGUGACCGGCGUUGCUGGCAACUAUUUCAAUUUGGCAGCCAAGUGGCUUUC ......(((((((.........))))...........((((((((((.((((.((((......))))..(((..(((((...)))))..))).....)))).))))))))))..)))... ( -36.20) >DroSim_CAF1 43759 120 + 1 UCAAUUAGCGGUCAGAUCCUUGGGCCAAUUCGACUAUUUUGGCUGCUUAAUUUUGCGUAAAAUCGCACAGGUGACCGGCGUUGCUGGCAACUAUUUCAAUUUGGCAGCCAAGUGGCUUUC ......(((((((.........))))...........((((((((((.((((.((((......))))..(((..(((((...)))))..))).....)))).))))))))))..)))... ( -36.20) >DroEre_CAF1 55233 119 + 1 UCAAUUAGCGGUCACAUCCUUGGGCCAAUUCGCCUAUUUUGGCUGCCUAAUUUUGCGUUAAAUCGUACAGGUGACCG-GGAUACUGGCAACUAUUUCAAUUUGGCAGCCAAGUGACUUUC .........((((((.....(((((......)))))..(((((((((.((((.((((......))))..(((..(((-(....))))..))).....)))).)))))))))))))))... ( -41.80) >DroYak_CAF1 55357 120 + 1 UCAAUUAGCGGUCACAUCCUUGGGCCAAUUCGCCUAUUUUGGCUGCCUAAUUUUGCGUUAAAUCGCACAGGUGACCGGGGAUACUGGCAACUAUUUCAAUUUGCCAGCCAAGUGACUUUC .........(((((((((((((((((((..........))))).((((.....((((......)))).))))..)))))))).(((((((..........)))))))....))))))... ( -40.90) >consensus UCAAUUAGCGGUCAGAUCCUUGGGCCAAUUCGCCUAUUUUGGCUGCUUAAUUUUGCGUAAAAUCGCACAGGUGACCGGCGAUACUGGCAACUAUUUCAAUUUGGCAGCCAAGUGGCUUUC (((......((((.........))))...........((((((((((.((((.((((......))))..(((..((((.....))))..))).....)))).)))))))))))))..... (-29.80 = -29.60 + -0.20)


| Location | 5,315,886 – 5,315,999 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.71 |
| Mean single sequence MFE | -36.32 |
| Consensus MFE | -31.48 |
| Energy contribution | -31.44 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5315886 113 + 22224390 GGCUGCUUAAUUUUGCGUAAAAUCGCACAGGUGACCGGCGAUACUGGCAACUAUUUCAAUUUGGCAGCCAAGUGGCUUUCCAGCACAUUAAACCCUGUCUGGCGUCUGC-------UCAC (((((((.((((.((((......))))..(((..((((.....))))..))).....)))).)))))))..(((((...((((.(((........))))))).....))-------.))) ( -35.00) >DroSec_CAF1 52316 113 + 1 GGCUGCUUAAUUUUGCGUAAAAUCGCACAGGUGACCGGCGUUGCUGGCAACUAUUUCAAUUUGGCAGCCAAGUGGCUUUCCAGCACAUUAAACCCUGUCUGGCGUCUGC-------CCAC (((((((.((((.((((......))))..(((..(((((...)))))..))).....)))).)))))))..(.(((...((((.(((........))))))).....))-------)).. ( -37.90) >DroSim_CAF1 43799 113 + 1 GGCUGCUUAAUUUUGCGUAAAAUCGCACAGGUGACCGGCGUUGCUGGCAACUAUUUCAAUUUGGCAGCCAAGUGGCUUUCCAGCACAUUAAACCCUGUCUGGCGUCUGC-------CCAC (((((((.((((.((((......))))..(((..(((((...)))))..))).....)))).)))))))..(.(((...((((.(((........))))))).....))-------)).. ( -37.90) >DroEre_CAF1 55273 112 + 1 GGCUGCCUAAUUUUGCGUUAAAUCGUACAGGUGACCG-GGAUACUGGCAACUAUUUCAAUUUGGCAGCCAAGUGACUUUCCAGCACAUUAAGCCCUGUCUGAAGUCUGC-------UCAC (((((((.((((.((((......))))..(((..(((-(....))))..))).....)))).))))))).((((((((..(((.(((........))))))))))).))-------)... ( -34.40) >DroYak_CAF1 55397 120 + 1 GGCUGCCUAAUUUUGCGUUAAAUCGCACAGGUGACCGGGGAUACUGGCAACUAUUUCAAUUUGCCAGCCAAGUGACUUUCCAGCACAUUAAACCCUGUCUGGAGUCUGCACUCCGCUCAC ((..((((.....((((......)))).))))..))((((...(((((((..........)))))))....(((((..(((((.(((........))))))))))).)).))))...... ( -36.40) >consensus GGCUGCUUAAUUUUGCGUAAAAUCGCACAGGUGACCGGCGAUACUGGCAACUAUUUCAAUUUGGCAGCCAAGUGGCUUUCCAGCACAUUAAACCCUGUCUGGCGUCUGC_______UCAC (((((((.((((.((((......))))..(((..((((.....))))..))).....)))).)))))))....(((...((((.(((........))))))).))).............. (-31.48 = -31.44 + -0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:23 2006