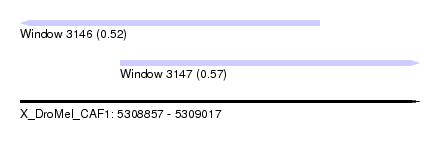
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,308,857 – 5,309,017 |
| Length | 160 |
| Max. P | 0.574431 |

| Location | 5,308,857 – 5,308,977 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.21 |
| Mean single sequence MFE | -27.64 |
| Consensus MFE | -22.46 |
| Energy contribution | -23.86 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5308857 120 - 22224390 AAUAAAAGGAUAUAUUCCUCUGGCCAGCGCUUUAAGCUCGCCUUUAUGUCUUUUGACAAAAGUCUUAGCGACUUAUUUGAGCCAGCCUGCAGCUACAGAUUUUAAUUACGGCUAUAAAUC ......((((.....)))).(((((...(((....((((.......((((....)))).(((((.....)))))....)))).)))(((......)))...........)))))...... ( -27.10) >DroSec_CAF1 45610 118 - 1 AAUAAAAGGAU--AUUCCUCUGGCCAGCGCUUUAAGCUCGCCUUUAUGUCUUUUGACAAAAGUCUUAGCGACUUAUUUGAGUCAGCCUGCAGCUACAGAUUUUAAUUACGGCUAUAAAUC ....(((((((--((......(((.(((.......))).)))...)))))))))....((((((((((((((((....))))).(....).)))).)))))))................. ( -28.70) >DroSim_CAF1 37126 118 - 1 AAUAAAAGGAU--AUUCCUCUGGCCAGCGCUUUAAGCUCGCCUUUAUGUCUUUUGACAAAAGUCUUAGCGACUUAUUUGAGUCAGCCUGCAGCUACAGAUUUUAAUUACGGCUAUAAAUC ....(((((((--((......(((.(((.......))).)))...)))))))))....((((((((((((((((....))))).(....).)))).)))))))................. ( -28.70) >DroEre_CAF1 48329 118 - 1 AAUAAAAGGAU--AUUCCUCUGGGCAGCGCUUUAAGCUCGCCUUUAUGUCUUUUGACAAAAGUCUUAGCGACUUAUUCGAGUCAGCCUGCAGCUACAGAUUUUAAUUACGGCUAUAAAUC ....(((((((--((......(((((((.......))).))))..)))))))))....((((((((((((((((....))))).(....).)))).)))))))................. ( -28.90) >DroYak_CAF1 48112 114 - 1 AAUAAAAGGAU--AUUGGUCUGGCCAGCGCUUUAAGCUCGCCUUUAUGUCUUUUGACAAAAGUCUUAGCGACUUAUUCGA----GCCUGCAGCUACAGAUUUUAAUUACGGCUAUAAAUC ........(((--...((((((...(((((.....(((((......((((....)))).(((((.....)))))...)))----))..)).))).))))))...)))............. ( -24.80) >consensus AAUAAAAGGAU__AUUCCUCUGGCCAGCGCUUUAAGCUCGCCUUUAUGUCUUUUGACAAAAGUCUUAGCGACUUAUUUGAGUCAGCCUGCAGCUACAGAUUUUAAUUACGGCUAUAAAUC ......((((.....))))..(((.(((.......))).)))....((((....))))((((((((((((((((....))))).(....).)))).)))))))................. (-22.46 = -23.86 + 1.40)


| Location | 5,308,897 – 5,309,017 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.12 |
| Mean single sequence MFE | -22.88 |
| Consensus MFE | -18.64 |
| Energy contribution | -19.48 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5308897 120 + 22224390 UCAAAUAAGUCGCUAAGACUUUUGUCAAAAGACAUAAAGGCGAGCUUAAAGCGCUGGCCAGAGGAAUAUAUCCUUUUAUUCCGGAAUUUAAUUAAGCCAAAUCCCCCUAAACACACACAC ......(((((.....))))).((((....))))....(((....(((((...((((..((((((.....))))))....))))..)))))....)))...................... ( -24.30) >DroSec_CAF1 45650 117 + 1 UCAAAUAAGUCGCUAAGACUUUUGUCAAAAGACAUAAAGGCGAGCUUAAAGCGCUGGCCAGAGGAAU--AUCCUUUUAUUCCGGAAUUUAAUUAAGCCAAAUCCCCCUAAACACACACA- ......(((((.....))))).((((....))))....(((.(((.......))).)))...(((((--(......))))))((.((((.........)))).))..............- ( -24.20) >DroSim_CAF1 37166 118 + 1 UCAAAUAAGUCGCUAAGACUUUUGUCAAAAGACAUAAAGGCGAGCUUAAAGCGCUGGCCAGAGGAAU--AUCCUUUUAUUCCGGAAUUUAAUUAAGCCAAAUCCCCCUAAACACACACAC ......(((((.....))))).((((....))))....(((.(((.......))).)))...(((((--(......))))))((.((((.........)))).))............... ( -24.20) >DroEre_CAF1 48369 118 + 1 UCGAAUAAGUCGCUAAGACUUUUGUCAAAAGACAUAAAGGCGAGCUUAAAGCGCUGCCCAGAGGAAU--AUCCUUUUAUUCCGGAAUUUAAUUAAGCCAAAUCCCCCUAAACACACACUC ......(((((.....))))).((((....))))....(((.(((.......))).......(((((--(......)))))).............)))...................... ( -21.00) >DroYak_CAF1 48148 118 + 1 UCGAAUAAGUCGCUAAGACUUUUGUCAAAAGACAUAAAGGCGAGCUUAAAGCGCUGGCCAGACCAAU--AUCCUUUUAUUCCGUAAUUUAAUUAGUCUAAAUCCUCCUAAACACACAUAC ..((..(((((.....)))))...))...((((.....(((.(((.......))).))).....(((--(......))))..............))))...................... ( -20.70) >consensus UCAAAUAAGUCGCUAAGACUUUUGUCAAAAGACAUAAAGGCGAGCUUAAAGCGCUGGCCAGAGGAAU__AUCCUUUUAUUCCGGAAUUUAAUUAAGCCAAAUCCCCCUAAACACACACAC ......(((((.....))))).((((....))))....(((....(((((...((((..((((((.....))))))....))))..)))))....)))...................... (-18.64 = -19.48 + 0.84)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:16 2006