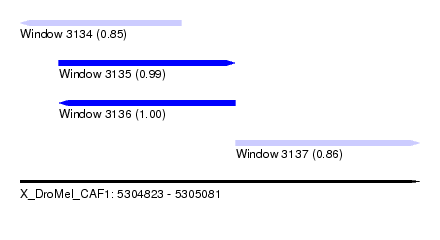
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,304,823 – 5,305,081 |
| Length | 258 |
| Max. P | 0.997991 |

| Location | 5,304,823 – 5,304,927 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.57 |
| Mean single sequence MFE | -34.84 |
| Consensus MFE | -23.32 |
| Energy contribution | -25.24 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5304823 104 - 22224390 AGAAUCCUAGGAAAAUCGAAAG-AAAAUCGAAGGAAAAUCCGUGCGUGUCUGGGUGGACUAAUGUGCAGACACUGCGAUUUGAUCGGUCAGUCUGGGAAUCCU----------GG----- .((.(((((((...(((((...-.(((((...((.....))..(((((((((.(..........).))))))).)))))))..)))))...))))))).))..----------..----- ( -32.30) >DroSec_CAF1 41527 107 - 1 AGAAUCCUAGGAAA------------AUCGAAGGAAAAUCCGUGCGUGUCUGGGUAGACUAAUGUGCAGACACUGCGAUUUGAUCGGUCAGUCUGGGAAUGCUUGGGUAUCGUGGGAAU- ....(((((.((..------------.((....))..(((((.((((..(..((..(((..((.(((((...))))).....))..)))..))..)..)))).))))).)).)))))..- ( -34.30) >DroSim_CAF1 32312 119 - 1 AGAAUCCUAGGAAAAUCGAAAGGAAAAUCGAAGGAAAAUCCGUGCGUGUCUGAGUAGACUAAUGUGCAGACACUGCGAUUUGAUCGGUCAGUCUGGGAAUGCCUGGGUAUCGUGGGAAU- ....(((((.((..(((((..(((...((....))...))).((((((((((..((......))..))))))).)))......)))))...((..((....))..))..)).)))))..- ( -36.10) >DroEre_CAF1 42873 117 - 1 GGAAUCGUAGGAAAAUCGGAGA-AAAAUCGAAGGAAAAUCCGUGCGUGUCUGGG-AGACUAAUGUGCAGACACUGCGAUUUGAUCGGUCAGUCUGGGAAUCCA-GGGCAUCCAGGGAAUC .(((((((((......((((..-....((....))...))))((((..(.(((.-...))))..))))....)))))))))....((....(((((....)))-))....))........ ( -34.10) >DroYak_CAF1 43625 117 - 1 AGACUCGUAGGAAAAUCGGAGA-AAAAUCGAAGGAAAUUGUGUGCGUGUCUGGG-AGACUAAUGUGCAGACACUGCGAUUUGAUCGGUCAGUCUGGGAAUCCU-UAGAAUCCUGGGAAUC .(((.((.....((((((.((.-....((....))...(((.((((..(.(((.-...))))..)))).))))).))))))...)))))..((..(((.((..-..)).)))..)).... ( -37.40) >consensus AGAAUCCUAGGAAAAUCGAAAA_AAAAUCGAAGGAAAAUCCGUGCGUGUCUGGGUAGACUAAUGUGCAGACACUGCGAUUUGAUCGGUCAGUCUGGGAAUCCU_GGGUAUCCUGGGAAU_ ....(((((((...(((((.....(((((...((.....))..(((((((((.(((......))).))))))).)))))))..)))))...)))))))...................... (-23.32 = -25.24 + 1.92)



| Location | 5,304,848 – 5,304,962 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.51 |
| Mean single sequence MFE | -20.23 |
| Consensus MFE | -12.94 |
| Energy contribution | -13.94 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5304848 114 + 22224390 AAUCGCAGUGUCUGCACAUUAGUCCACCCAGACACGCACGGAUUUUCCUUCGAUUUU-CUUUCGAUUUUCCUAGGAUUCUCCAUUC-----CAACAUUCCCCCCUGAUGAACUGCGACUG ..((((((((((((..............)))))))....(((...((((..((...(-(....))...))..))))...)))....-----.....................)))))... ( -25.24) >DroSec_CAF1 41566 99 + 1 AAUCGCAGUGUCUGCACAUUAGUCUACCCAGACACGCACGGAUUUUCCUUCGAU------------UUUCCUAGGAUUCUCCCUAC-----CAACUUACCCC----AUGAGCUGCGACUG ..((((((((((((..............)))))))((..(((...((....)).------------..)))((((......)))).-----...........----....)))))))... ( -23.24) >DroSim_CAF1 32351 111 + 1 AAUCGCAGUGUCUGCACAUUAGUCUACUCAGACACGCACGGAUUUUCCUUCGAUUUUCCUUUCGAUUUUCCUAGGAUUCUCCCUAC-----CAACUUACCCC----AUCAACUGCGACUG ..((((((((((((..............)))))))....(((...((((((((........)))).......))))...)))....-----...........----......)))))... ( -24.05) >DroEre_CAF1 42912 96 + 1 AAUCGCAGUGUCUGCACAUUAGUCU-CCCAGACACGCACGGAUUUUCCUUCGAUUUU-UCUCCGAUUUUCCUACGAUUCCCCUCCUCCCCCCGCACU----------------------G ((((((.(((((((.((....))..-..))))))))).((((...............-..))))..........))))...................----------------------. ( -15.53) >DroYak_CAF1 43664 91 + 1 AAUCGCAGUGUCUGCACAUUAGUCU-CCCAGACACGCACACAAUUUCCUUCGAUUUU-UCUCCGAUUUUCCUACGAGUCUCCUUCC-----CAACUU----------------------A ....((.(((((((.((....))..-..)))))))))....................-.....(((((......))))).......-----......----------------------. ( -13.10) >consensus AAUCGCAGUGUCUGCACAUUAGUCUACCCAGACACGCACGGAUUUUCCUUCGAUUUU_CCUCCGAUUUUCCUAGGAUUCUCCCUAC_____CAACUU_CCCC____AU_A_CUGCGACUG ....((.(((((((..............)))))))))..(((...((((..((...............))..))))...)))...................................... (-12.94 = -13.94 + 1.00)

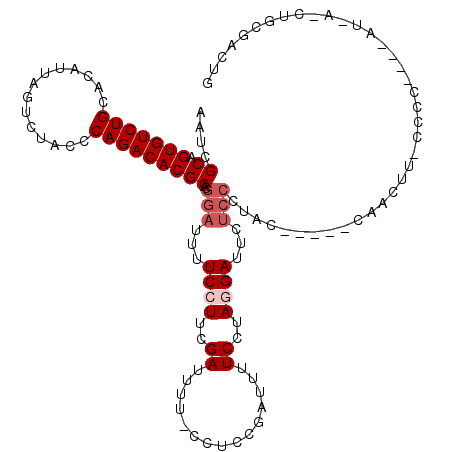

| Location | 5,304,848 – 5,304,962 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.51 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -15.78 |
| Energy contribution | -15.90 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.997991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5304848 114 - 22224390 CAGUCGCAGUUCAUCAGGGGGGAAUGUUG-----GAAUGGAGAAUCCUAGGAAAAUCGAAAG-AAAAUCGAAGGAAAAUCCGUGCGUGUCUGGGUGGACUAAUGUGCAGACACUGCGAUU .(((((((((((.(((((.(.(.......-----..(((((...((((..((...((....)-)...))..))))...))))).).).))))).((.((....)).)))).))))))))) ( -33.80) >DroSec_CAF1 41566 99 - 1 CAGUCGCAGCUCAU----GGGGUAAGUUG-----GUAGGGAGAAUCCUAGGAAA------------AUCGAAGGAAAAUCCGUGCGUGUCUGGGUAGACUAAUGUGCAGACACUGCGAUU .((((((((((...----..)))......-----(((.(((...((((..(...------------..)..))))...))).)))(((((((.(((......))).)))))))))))))) ( -31.80) >DroSim_CAF1 32351 111 - 1 CAGUCGCAGUUGAU----GGGGUAAGUUG-----GUAGGGAGAAUCCUAGGAAAAUCGAAAGGAAAAUCGAAGGAAAAUCCGUGCGUGUCUGAGUAGACUAAUGUGCAGACACUGCGAUU .(((((((......----...........-----(((.(((...((((..((...((....))....))..))))...))).)))(((((((..((......))..)))))))))))))) ( -34.10) >DroEre_CAF1 42912 96 - 1 C----------------------AGUGCGGGGGGAGGAGGGGAAUCGUAGGAAAAUCGGAGA-AAAAUCGAAGGAAAAUCCGUGCGUGUCUGGG-AGACUAAUGUGCAGACACUGCGAUU .----------------------...................((((((((......((((..-....((....))...))))((((..(.(((.-...))))..))))....)))))))) ( -24.50) >DroYak_CAF1 43664 91 - 1 U----------------------AAGUUG-----GGAAGGAGACUCGUAGGAAAAUCGGAGA-AAAAUCGAAGGAAAUUGUGUGCGUGUCUGGG-AGACUAAUGUGCAGACACUGCGAUU .----------------------...(((-----.((.(....))).)))...(((((.((.-....((....))...(((.((((..(.(((.-...))))..)))).))))).))))) ( -25.70) >consensus CAGUCGCAG_U_AU____GGGG_AAGUUG_____GGAAGGAGAAUCCUAGGAAAAUCGAAAA_AAAAUCGAAGGAAAAUCCGUGCGUGUCUGGGUAGACUAAUGUGCAGACACUGCGAUU ......................................(((...((....))...............((....))...))).((((((((((.(((......))).))))))).)))... (-15.78 = -15.90 + 0.12)

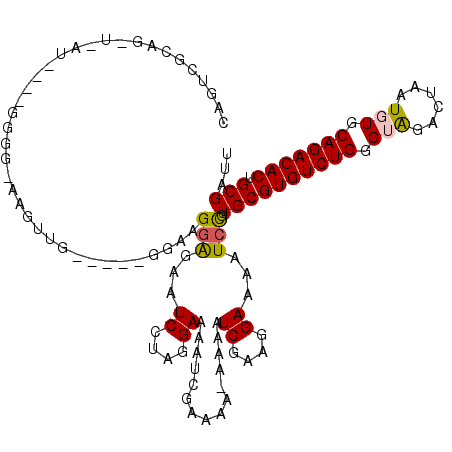

| Location | 5,304,962 – 5,305,081 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.32 |
| Mean single sequence MFE | -39.50 |
| Consensus MFE | -28.76 |
| Energy contribution | -29.84 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5304962 119 + 22224390 CCACUGCCACUGGCACUUGCCUUGCGCUUUUCCCUGGCUCCUGUCUAAAUGUCCAUCUGCCAGUUUCAAUUUGAAACAAUGUCGCCCGGG-GCCAGAGAAGUGCAAUUACUUUCAUGUUU ...........(((....)))(((((((((((..(((((((.(.(.....).).........(((((.....)))))..........)))-))))))))))))))).............. ( -33.60) >DroSec_CAF1 41665 119 + 1 CCACUGCCACUGGCACUUGCCUUGCGCUUUUCCUUGGCUCCUGUCCAGAUGUCCAUCUGCCAGUUUUAAUUUGAAACAAUGUCGCCGGGG-GCCAGAGAAGUGCAAUUACUUUCAUGUUU ...........(((....)))(((((((((((..(((((((((..(((((....)))))...(((((.....)))))........)))))-))))))))))))))).............. ( -40.20) >DroSim_CAF1 32462 119 + 1 CCACUGCCACUGGCACUUGCCUUGCGCUUUUCCUUGGCUCCUGUCCAGAUGUCCAUCUGCCAGUUUCAAUUUGAAACAAUGUCGCCGGGG-GCCAGAGAAGUGCAAUUACUUUCAUGUUU ...........(((....)))(((((((((((..(((((((((..(((((....)))))...(((((.....)))))........)))))-))))))))))))))).............. ( -42.60) >DroEre_CAF1 43008 120 + 1 CGACUGCCACUGGCACUCUCCUUGCGCUUUUCCUUGGCUCCCGUCUAGAUGUCCAUCUGCCAGUUUCCAUUUGAAACAAUGUCGCCCGGGGGCCAGAGAAGUGCAAUUACUUUCAUGUUU .((.((((...)))).))...(((((((((((..(((((((((..(((((....)))))...(((((.....))))).........)))))))))))))))))))).............. ( -43.30) >DroYak_CAF1 43755 120 + 1 CCACUGCCAGUGGCACUUGCCAUGCGCUUUUCCUCGGCUGCUGUCUGAAUGUCCAUCUGCCAGUUUCAAUUUGAAACAAUGUCGCCCGGGGCCGAGAGAAGUGCAAUUACUUUCAUGUUU .........(((((....)))))(((((((((.((((((.(((..(((..((......))..(((((.....)))))....)))..))))))))))))))))))................ ( -37.80) >consensus CCACUGCCACUGGCACUUGCCUUGCGCUUUUCCUUGGCUCCUGUCUAGAUGUCCAUCUGCCAGUUUCAAUUUGAAACAAUGUCGCCCGGG_GCCAGAGAAGUGCAAUUACUUUCAUGUUU ....((((...))))......(((((((((((..((((.((((..(((((....)))))...(((((.....))))).........)))).))))))))))))))).............. (-28.76 = -29.84 + 1.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:06 2006