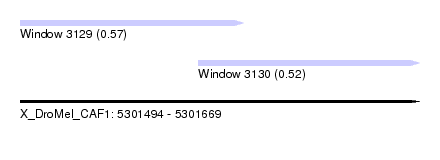
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,301,494 – 5,301,669 |
| Length | 175 |
| Max. P | 0.572819 |

| Location | 5,301,494 – 5,301,592 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 85.13 |
| Mean single sequence MFE | -27.35 |
| Consensus MFE | -19.33 |
| Energy contribution | -19.32 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5301494 98 + 22224390 ACAUCGCACAGCCAAGUGGAAGUGCGC-UGUUAAAUGAGCGUGUUGCCCGCGGAAAGUCCAACAGCAAAACUAGCAACUUGCAACUUGCAACAUGCCAA (((.(((((..((....))..))))).-)))....((.(((((((((....((.....))....((((..........)))).....))))))))))). ( -31.40) >DroSec_CAF1 38309 87 + 1 ACAUCGCACAGCCAAGUGCAAG--CGC-UGUUAAAUGAGCGUGUUGCCCGCGGAAGGUCCA---------CUUGCAACUUGCUACUUGCAACAUGCCAC .....(((....((((((((((--(((-(........)))))(((((..(.((.....)).---------)..))))))))).))))).....)))... ( -25.40) >DroSim_CAF1 29145 87 + 1 ACAUCGCACAGCCAAGUGGAAG--CGC-UGUUAAAUGAGCGUGUUGCCCGCGGAAGGUCCA---------CUUGCAACUUGCUACUUGCAACAUGCCAC .....(((..((.(((((((.(--(((-(........)))))........(....).))))---------)))))...((((.....))))..)))... ( -24.80) >DroYak_CAF1 40128 82 + 1 ----CGCACAUCCAAGUGGAUG--CGUUUGUUAAAUGAGCGUGUUGC-CGCGGAAGGUCCA---------CUGGCAACUUGCAGCUUGCAACAUGCCA- ----.((.(((((....)))))--.))........((.(((((((((-.((((.....)).---------...((.....)).))..)))))))))))- ( -27.80) >consensus ACAUCGCACAGCCAAGUGGAAG__CGC_UGUUAAAUGAGCGUGUUGCCCGCGGAAGGUCCA_________CUUGCAACUUGCAACUUGCAACAUGCCAC ......(((......))).................((.(((((((((....((.....)).............((.....)).....))))))))))). (-19.33 = -19.32 + -0.00)



| Location | 5,301,572 – 5,301,669 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.58 |
| Mean single sequence MFE | -34.16 |
| Consensus MFE | -23.72 |
| Energy contribution | -23.16 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5301572 97 + 22224390 -------------UGCAACUUGCAACAUGCCAA--------CGCCGUGUGGCACUUUGUUGGGCAAAAGGACCUGAUUGCCAGCAAGGAUAC--CAACGAUGCUGCUUGUAAAAGCCGAA -------------(((((...(((.(.(((((.--------.......)))))(((((((((.(((.((...))..))))))))))))....--....).)))...)))))......... ( -24.80) >DroSec_CAF1 38376 96 + 1 -------------UGCUACUUGCAACAUGCCAC--------CGCCGUGUGGCACUUUGUUG-GCGAAAGGAGCUGAUUGCCAGCAAGGAUAC--CAGCCAUGCUGCUUGUAAAAGCCGAA -------------.(((..((((((..((((((--------......))))))((((((((-((((.((...))..))))))))))))....--((((...)))).)))))).))).... ( -32.30) >DroSim_CAF1 29212 96 + 1 -------------UGCUACUUGCAACAUGCCAC--------CGCCGUGUGGCACUUUGUUG-GCGAAAGGAGCUGAUUGCCAGCAAGGAUAC--CAGCCAUGCUGCUUGUAAAAGCCGAA -------------.(((..((((((..((((((--------......))))))((((((((-((((.((...))..))))))))))))....--((((...)))).)))))).))).... ( -32.30) >DroEre_CAF1 39762 113 + 1 GCCGCCUGCAGCUUGCAGCUUGCAACAUGCCACCGCUACCCCGCCGUGUGGCACUUUGUUG-GCAAGAGGACCUGAUUGCCAGCAAGGAUGCCG------UGCUGCUUGUAAAAGCCGAA ..((.((...((..(((((..(((...(((((((((......).)).))))))((((((((-((((..(....)..)))))))))))).)))..------.)))))..))...)).)).. ( -45.10) >DroYak_CAF1 40191 97 + 1 -------------UGCAGCUUGCAACAUGCCA---------CGCGGUGUUGCACUUUGUUG-GCAAGAGGAGCUGAUUGCCAGCAAGGAUACUGCAAGGAUGCUGCUUGUAAAAGCCGAA -------------(((((..((((((((((..---------.)).))))))))((((((((-((((..(....)..))))))))))))...)))))......(.((((....)))).).. ( -36.30) >consensus _____________UGCAACUUGCAACAUGCCAC________CGCCGUGUGGCACUUUGUUG_GCAAAAGGAGCUGAUUGCCAGCAAGGAUAC__CAACCAUGCUGCUUGUAAAAGCCGAA ..............((...((((((((.((............(((....))).((((((((.((((..(....)..)))))))))))).............)))).))))))..)).... (-23.72 = -23.16 + -0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:00 2006