| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 660,740 – 660,852 |
| Length | 112 |
| Max. P | 0.759235 |

| Location | 660,740 – 660,852 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.26 |
| Mean single sequence MFE | -39.07 |
| Consensus MFE | -24.49 |
| Energy contribution | -26.07 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
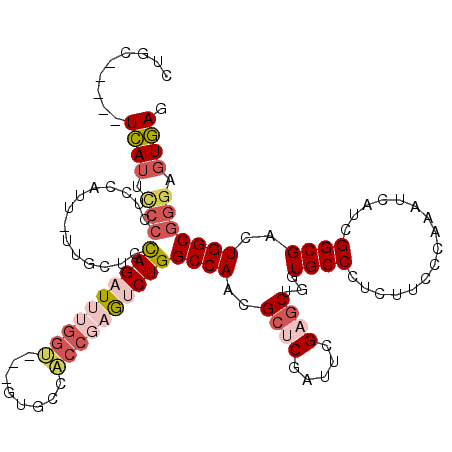
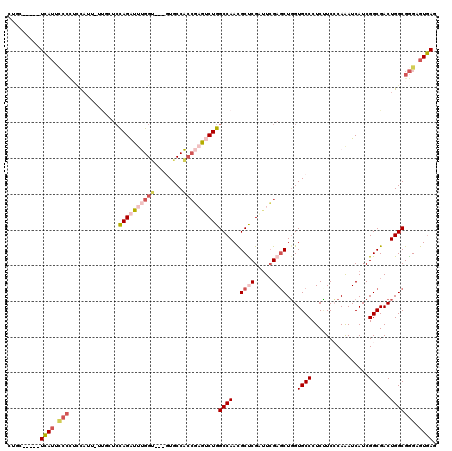
>X_DroMel_CAF1 660740 112 - 22224390 CGGC-----UCAUUUCCCUCCAUUUUUGCUCUAGAUUUGGU---GUGCCACCGAGUCUGGCCAACGCUCGAUUCGAGCUGGUGCCCUCUUCCCAAAUCAUCGGCGACUGGCGGGUGUGAG ...(-----((((..(((.(((...(((((...(((((((.---(..((((((((((.(((....))).)))))).).)))..).......)))))))...))))).))).))).))))) ( -42.10) >DroSec_CAF1 10758 112 - 1 CUGC-----UCAUUCCCCACCAUUUUUGCUCUAGAUUUGGU---GUGCCACCGAGUCUGGCCAACGCUCGAUUCGAGCUGGUGCCCUCUUCCCAAAUCAUCGGCGACUGGCGGGUGUGAG ...(-----((((..(((.(((...(((((...(((((((.---(..((((((((((.(((....))).)))))).).)))..).......)))))))...))))).))).))).))))) ( -42.00) >DroSim_CAF1 6422 112 - 1 CUGC-----UCAUUCCCCUCCAUUUUUGCUCUAGAUUUGGU---GUGCCACCGAGUCUGGCCAACGCUCGAUUCGAGCUGGUGCCCUCUUCCCAAAUCAUCGGCGACUGGCGGGUGUGAG ...(-----((((..(((.(((...(((((...(((((((.---(..((((((((((.(((....))).)))))).).)))..).......)))))))...))))).))).))).))))) ( -41.60) >DroEre_CAF1 6148 111 - 1 CCGC-----UCAUUCCGCUGUAUC-UUGCUCCAGAUUUGGU---GUGCCACCGAGUCUGGCCAACGCCCGCUUCGAGCUGGUGCCCUCCUCACAAAUCAUCGGCGACUGGCGGGAGUGAU .(((-----((.....((......-..)).(((((((((((---(...)))))))))))).......(((((....(((((((..............)))))))....)))))))))).. ( -41.94) >DroAna_CAF1 18793 109 - 1 -UUC-----UUAUUACCAAUC-----CCCGACAGAUACGGCAACUUGCCGCCUCACCUGGCCAAUGCCCGCUUCGAACUGGUGCCCUCCUCCCAAAUCAUCGGCGACUGGCGGGAGUAAU -...-----............-----((((.(((....((((..(((..(((......))))))))))((((..((..(((..........)))..))...)))).))).))))...... ( -26.80) >DroPer_CAF1 13123 111 - 1 -GUCUUAUAUCAGUCACUUCCAUU-----UCCAGGUUCGGA---AUGCCGGACCAGCUGGCCAACGCUCGCCUGGAGCUCGUGCCCUCGUCGCAGAUCAUCGGCGACUGGCGAGAGUGAA -............(((((..(..(-----(((((((((((.---...))))))).((.(((....))).)).))))).....(((...(((((.(.....).))))).))))..))))). ( -40.00) >consensus CUGC_____UCAUUCCCCUCCAUU_UUGCUCCAGAUUUGGU___GUGCCACCGAGUCUGGCCAACGCUCGAUUCGAGCUGGUGCCCUCUUCCCAAAUCAUCGGCGACUGGCGGGAGUGAG .........((((.(((..............((((((((((........))))))))))((((..((((.....))))...((((................))))..))))))).)))). (-24.49 = -26.07 + 1.59)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:38 2006