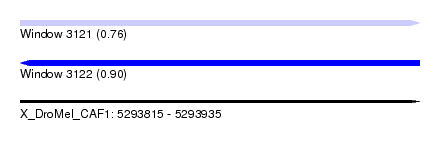
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,293,815 – 5,293,935 |
| Length | 120 |
| Max. P | 0.903081 |

| Location | 5,293,815 – 5,293,935 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.57 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -17.24 |
| Energy contribution | -17.90 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
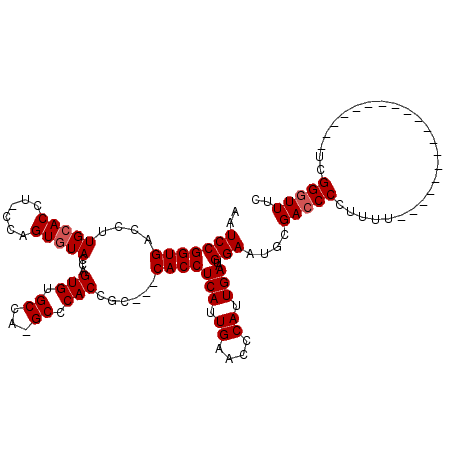
>X_DroMel_CAF1 5293815 120 + 22224390 AAUCCGGUGACCUUGCACCUUCCAGUUUAGCAGUGUGCCAAGCCCACCUCCUGCACCUCAUUGAAUCCAUUGAAUGGAAUGCGACCCCUUUUCUUUUUUCUAUUUUUUUUCUGGGUUUUU (((((((((..(((((((((.(.......).)).))).))))..))))...((((..(((((.((....)).)))))..)))).............................)))))... ( -23.10) >DroSec_CAF1 30964 94 + 1 AAUCCGGUGACCUUGCACA---CUGUGUACCAGUGUGCCA-GCCCACCGC---CACCUCAUUGAACCCAUUGAAUGGAAUGCGACCCCUUUU-------------------UCGGGUUCC ....(((((..((.(((((---(((.....)))))))).)-)..))))).---.........((((((...(((.((........)).))).-------------------..)))))). ( -33.60) >DroSim_CAF1 21748 96 + 1 AAUCCGGUGACCUUGCACCU-CCAGUGUACCAGUGUGCCA-GCCCACCGC---CACCUCAUUGAACCCAUUGAAUGGAAUGCGACCCCUUUU-------------------UCGGGUUUC ....(((((..((.((((((-..........)).)))).)-)..)))))(---((..(((.((....)).))).))).....(((((.....-------------------..))))).. ( -23.50) >consensus AAUCCGGUGACCUUGCACCU_CCAGUGUACCAGUGUGCCA_GCCCACCGC___CACCUCAUUGAACCCAUUGAAUGGAAUGCGACCCCUUUU___________________UCGGGUUUC ..(((((((....(((((......)))))...(((.((...)).)))......))))(((.((....)).)))..)))....(((((..........................))))).. (-17.24 = -17.90 + 0.67)


| Location | 5,293,815 – 5,293,935 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.57 |
| Mean single sequence MFE | -34.65 |
| Consensus MFE | -20.08 |
| Energy contribution | -19.86 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
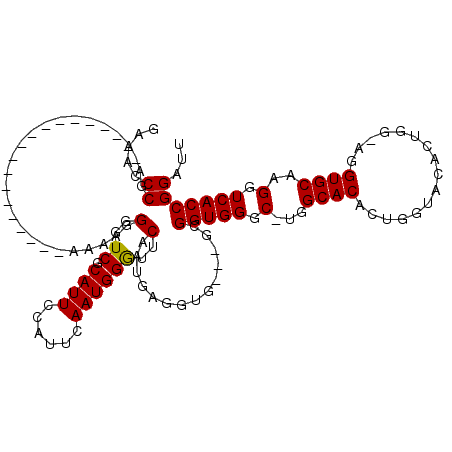
>X_DroMel_CAF1 5293815 120 - 22224390 AAAAACCCAGAAAAAAAAUAGAAAAAAGAAAAGGGGUCGCAUUCCAUUCAAUGGAUUCAAUGAGGUGCAGGAGGUGGGCUUGGCACACUGCUAAACUGGAAGGUGCAAGGUCACCGGAUU .....(((........................)))(((((((..((((.((....)).))))..))))....(((((.((((.(((.((.((.....)).))))))))).))))).))). ( -29.76) >DroSec_CAF1 30964 94 - 1 GGAACCCGA-------------------AAAAGGGGUCGCAUUCCAUUCAAUGGGUUCAAUGAGGUG---GCGGUGGGC-UGGCACACUGGUACACAG---UGUGCAAGGUCACCGGAUU .(((((((.-------------------.....(((......)))......))))))).........---.((((((.(-(.((((((((.....)))---))))).)).)))))).... ( -41.40) >DroSim_CAF1 21748 96 - 1 GAAACCCGA-------------------AAAAGGGGUCGCAUUCCAUUCAAUGGGUUCAAUGAGGUG---GCGGUGGGC-UGGCACACUGGUACACUGG-AGGUGCAAGGUCACCGGAUU ....(((..-------------------....)))(((.....(((((((.((....)).))).)))---)((((((.(-(.((((.(..(....)..)-..)))).)).))))))))). ( -32.80) >consensus GAAACCCGA___________________AAAAGGGGUCGCAUUCCAUUCAAUGGGUUCAAUGAGGUG___GCGGUGGGC_UGGCACACUGGUACACUGG_AGGUGCAAGGUCACCGGAUU .....((..........................(..((.((((......))))))..)..............(((((.(...((((................))))..).)))))))... (-20.08 = -19.86 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:53 2006