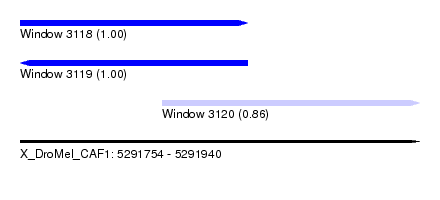
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,291,754 – 5,291,940 |
| Length | 186 |
| Max. P | 0.997631 |

| Location | 5,291,754 – 5,291,860 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.57 |
| Mean single sequence MFE | -40.82 |
| Consensus MFE | -32.98 |
| Energy contribution | -32.90 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.90 |
| SVM RNA-class probability | 0.997631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
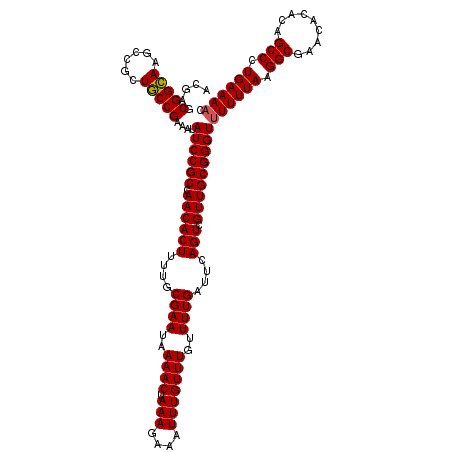
>X_DroMel_CAF1 5291754 106 + 22224390 AAAA----GCGGAUAUCCAGCAGCUGGGAACACUCCAUCGAGGUUUUUCC----------CGACGCUUUUCCGAACGCCAACGAGCGGCAAGCCGCUGCCGAAAUAUCCGCCAACACUUU ....----((((((((...(((((((((((..(((....)))....))))----------))..((((..(((..((....))..))).)))).)))))....))))))))......... ( -37.40) >DroSec_CAF1 28976 106 + 1 AAAA----GCGGAUAUCCAGAAACUGGGAACACUCCAUCGAGGUUUUUCC----------CGACGCUUCUCCGAGCGCCAACGAACGGAAAGCCGCUUCCGAAAUAUCCGCCAACACUUU ....----((((((((...(((.(((((((..(((....)))....))))----------))..((((.((((..((....))..)))))))).).)))....))))))))......... ( -31.10) >DroSim_CAF1 19782 106 + 1 AAAA----GCGGAUAUCCAGCAGCUGGGAACACUCCAUCGAGGUUUUUCC----------CGACGCUUUUCCGAGCGCCAACGAGCGGCAAGCCGCUGCCGAAAUAUCCGCCAACACUUU ....----((((((((...(((((((((((..(((....)))....))))----------))..((((..(((..((....))..))).)))).)))))....))))))))......... ( -38.80) >DroEre_CAF1 30138 118 + 1 AAACG--GGUGGAUAUCCAGCAGCUGGGAACACUCCAUCGAGGUUUUUCCGAGGCUUUUCCGAGGCUUUUCCGAGCGGCAACGAGCGGCAAGCCGCUGCCGAAAUAUCCGCCAACACUUU .....--(((((((((...(((((((((((....((.(((.((....)))))))..)))))).(((((..(((..((....))..))).))))))))))....)))))))))........ ( -47.90) >DroYak_CAF1 30099 109 + 1 AGUAGGCGGCGGAUAUCCAGCAGCUGGGAACACUCCAUCGAGGUUUUUCCGAGG-----------CUUUUCCGAGCGGCAACGAGCGGCAAGCCGCUGCCGAAAUAUCCGCCAACACUUU (((.(..(((((((((...(((((((((((..(((....)))...)))))).((-----------(((..(((..((....))..))).))))))))))....)))))))))..)))).. ( -48.90) >consensus AAAA____GCGGAUAUCCAGCAGCUGGGAACACUCCAUCGAGGUUUUUCC__________CGACGCUUUUCCGAGCGCCAACGAGCGGCAAGCCGCUGCCGAAAUAUCCGCCAACACUUU ........((((((((...(((((.(((((..(((....)))...)))))..............((((..(((..((....))..))).)))).)))))....))))))))......... (-32.98 = -32.90 + -0.08)

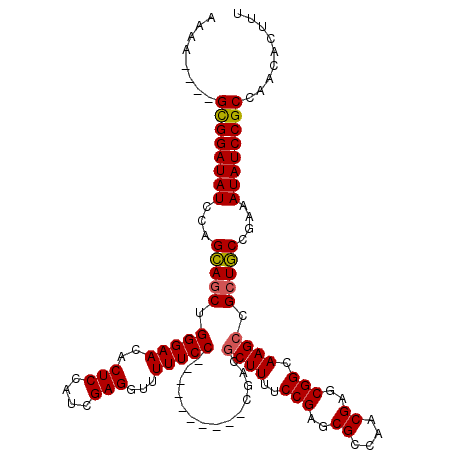
| Location | 5,291,754 – 5,291,860 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.57 |
| Mean single sequence MFE | -46.66 |
| Consensus MFE | -36.82 |
| Energy contribution | -36.74 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.996778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5291754 106 - 22224390 AAAGUGUUGGCGGAUAUUUCGGCAGCGGCUUGCCGCUCGUUGGCGUUCGGAAAAGCGUCG----------GGAAAAACCUCGAUGGAGUGUUCCCAGCUGCUGGAUAUCCGC----UUUU ........((((((((((.(((((((.((((.(((..((....))..)))..))))...(----------((((....(((....)))..))))).))))))))))))))))----)... ( -47.50) >DroSec_CAF1 28976 106 - 1 AAAGUGUUGGCGGAUAUUUCGGAAGCGGCUUUCCGUUCGUUGGCGCUCGGAGAAGCGUCG----------GGAAAAACCUCGAUGGAGUGUUCCCAGUUUCUGGAUAUCCGC----UUUU ........((((((((((.(((((((.((((((((..((....))..))))).)))...(----------((((....(((....)))..))))).))))))))))))))))----)... ( -45.00) >DroSim_CAF1 19782 106 - 1 AAAGUGUUGGCGGAUAUUUCGGCAGCGGCUUGCCGCUCGUUGGCGCUCGGAAAAGCGUCG----------GGAAAAACCUCGAUGGAGUGUUCCCAGCUGCUGGAUAUCCGC----UUUU ........((((((((((.(((((((((....)).......((((((......))))))(----------((((....(((....)))..))))).))))))))))))))))----)... ( -47.60) >DroEre_CAF1 30138 118 - 1 AAAGUGUUGGCGGAUAUUUCGGCAGCGGCUUGCCGCUCGUUGCCGCUCGGAAAAGCCUCGGAAAAGCCUCGGAAAAACCUCGAUGGAGUGUUCCCAGCUGCUGGAUAUCCACC--CGUUU ........((.(((((((.((((((((((((.(((..((....))..)))..)))))..((((...((((((.......)))).))....))))..)))))))))))))).))--..... ( -44.60) >DroYak_CAF1 30099 109 - 1 AAAGUGUUGGCGGAUAUUUCGGCAGCGGCUUGCCGCUCGUUGCCGCUCGGAAAAG-----------CCUCGGAAAAACCUCGAUGGAGUGUUCCCAGCUGCUGGAUAUCCGCCGCCUACU ..((((.(((((((((((.((((((((((((.(((..((....))..)))..)))-----------))..((((....(((....)))..))))..))))))))))))))))))..)))) ( -48.60) >consensus AAAGUGUUGGCGGAUAUUUCGGCAGCGGCUUGCCGCUCGUUGGCGCUCGGAAAAGCGUCG__________GGAAAAACCUCGAUGGAGUGUUCCCAGCUGCUGGAUAUCCGC____UUUU .........(((((((((.(((((((.((((.(((..((....))..)))..))))..............((((....(((....)))..))))..))))))))))))))))........ (-36.82 = -36.74 + -0.08)

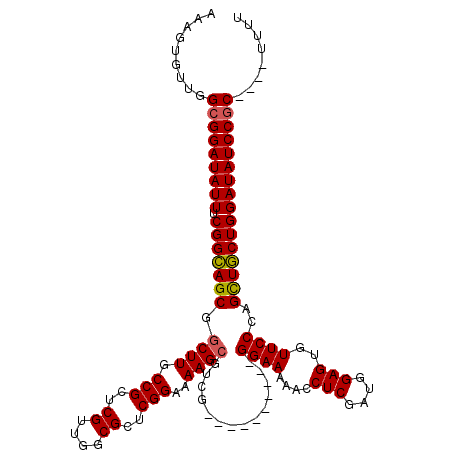

| Location | 5,291,820 – 5,291,940 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -32.72 |
| Consensus MFE | -27.88 |
| Energy contribution | -29.76 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5291820 120 + 22224390 ACGAGCGGCAAGCCGCUGCCGAAAUAUCCGCCAACACUUUUGCGAAUAAAACUAAAGAAAUUUGUUUGUUUUGAUUCAGUCGUUGCGGGUUUUUUAAGGCGAACACACAGCCCUGAAAAC .((.(((((.....)))))))....((((((.((((((....((((..((((.(((....)))))))..))))....))).)))))))))((((((.(((.........))).)))))). ( -33.00) >DroSec_CAF1 29042 120 + 1 ACGAACGGAAAGCCGCUUCCGAAAUAUCCGCCAACACUUUUGCGAAUAAAACUAAAGAAAUUUGUUUGUUUUGAUUCAGUCGUUGCGGGUUUUUUAAGGCGAACACACAGCCCUGAAAAC .....(((((......)))))....((((((.((((((....((((..((((.(((....)))))))..))))....))).)))))))))((((((.(((.........))).)))))). ( -29.00) >DroSim_CAF1 19848 120 + 1 ACGAGCGGCAAGCCGCUGCCGAAAUAUCCGCCAACACUUUUGCGAAUAAAACUAAAGAAAUUUGUUUGUUUUGAUUCAGUCGUUGCGGGUUUUUUAAGGCGAACACACAGCCCUGAAAAC .((.(((((.....)))))))....((((((.((((((....((((..((((.(((....)))))))..))))....))).)))))))))((((((.(((.........))).)))))). ( -33.00) >DroEre_CAF1 30216 119 + 1 ACGAGCGGCAAGCCGCUGCCGAAAUAUCCGCCAACACUUUUUCGAAUAAAACUAAAGAAAUUUGUUUGUUUUGAUUCAGUCGUUGCGGGUUUUUUAAGGCGAACACACAGCCCUGAAA-G .((.(((((.....)))))))....((((((.((((((...(((((..((((.(((....)))))))..)))))...))).))))))))).(((((.(((.........))).)))))-. ( -34.00) >DroYak_CAF1 30168 120 + 1 ACGAGCGGCAAGCCGCUGCCGAAAUAUCCGCCAACACUUUUUCGAAUAAAACUAAAGAAAUUUGUUUGUUUUGAUUCAGUCGUUGCGGGUUUUUUAAGGCGAACACACAGCCCUGAAAAC .((.(((((.....)))))))....((((((.((((((...(((((..((((.(((....)))))))..)))))...))).)))))))))((((((.(((.........))).)))))). ( -34.60) >consensus ACGAGCGGCAAGCCGCUGCCGAAAUAUCCGCCAACACUUUUGCGAAUAAAACUAAAGAAAUUUGUUUGUUUUGAUUCAGUCGUUGCGGGUUUUUUAAGGCGAACACACAGCCCUGAAAAC .....(((((......)))))....((((((.((((((....((((..((((.(((....)))))))..))))....))).)))))))))((((((.(((.........))).)))))). (-27.88 = -29.76 + 1.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:51 2006