| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,289,854 – 5,290,029 |
| Length | 175 |
| Max. P | 0.849117 |

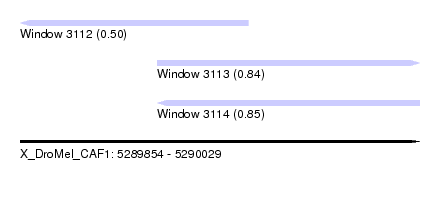
| Location | 5,289,854 – 5,289,954 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 83.89 |
| Mean single sequence MFE | -34.33 |
| Consensus MFE | -26.29 |
| Energy contribution | -27.35 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.08 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5289854 100 - 22224390 UAAUUGCAUUUUUGCGCUCCUGCCUGGGGCGUGGGCAGAACGGUCCGGACCGGAAGUCCUGUCCAGGACA---------ACGAAAGCCACAGAAACAAAAACGAGCUGA ..........((..((((((.....))))))..))(((..((((((((((.((....)).)))).)))).---------.(....)...............))..))). ( -34.70) >DroSec_CAF1 27192 100 - 1 UAAUUGCAUUUUUGCGCUCCUGCCUGCGGCGUGGGCAGAACGGUCCGGACCGGAAGUCCUCCUCAGGACA---------CCGAAUGGCGCAGAAACAAAAAGGAGCUGA .....(((....)))((((((..(((((.(((((((......))))....(((..(((((....))))).---------))).))).)))))........))))))... ( -37.50) >DroSim_CAF1 18012 100 - 1 UAAUUGCAUUUUUGCGCUCCUGCCUGCGGCGUGUGCAGAAUGGUCCGGACCGGAAGUCCUCCUCAGGACA---------CCGAAAGGCGCAGAAACAAAAAGGAGCUGA .....(((....)))(((((((((...))).(((((....(((((((...)))).(((((....))))).---------)))....))))).........))))))... ( -34.60) >DroEre_CAF1 28423 105 - 1 UAAUUGCAUUUUUGCGCUCU-UCCUGGGGCACGGGCAGAACAGA--AGUCCGGAAGUCCUU-CCAGGACACCACGCGCGCCGAAAGCCACAGAAACAAAAAGGAGCUGA .....((.((((.(((((((-.((((.....)))).))).....--.((((((((....))-)).)))).......)))).))))))..(((...(.....)...))). ( -30.50) >consensus UAAUUGCAUUUUUGCGCUCCUGCCUGCGGCGUGGGCAGAACGGUCCGGACCGGAAGUCCUCCCCAGGACA_________CCGAAAGCCACAGAAACAAAAAGGAGCUGA .....(((....)))((((((..(((((((..........((((....))))...(((((....)))))................)))))))........))))))... (-26.29 = -27.35 + 1.06)

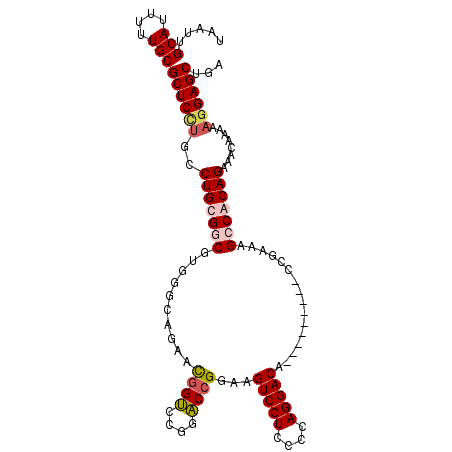

| Location | 5,289,914 – 5,290,029 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.35 |
| Mean single sequence MFE | -39.88 |
| Consensus MFE | -25.42 |
| Energy contribution | -27.54 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.836452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5289914 115 + 22224390 UUCUGCCCACGCCCCAGGCAGGAGCGCAAAAAUGCAAUUAUAAUCCCAG-----UGGCAGCAGGCGGCAACUGGCAGUGCAACUGCAUUGAAAAUUGCGCCUGCAAGUGCAACUUUGCAU (((((((.........)))))))..(((((..((((........((...-----.))..(((((((.(((.(..((((((....))))))..).))))))))))...))))..))))).. ( -43.40) >DroSec_CAF1 27252 115 + 1 UUCUGCCCACGCCGCAGGCAGGAGCGCAAAAAUGCAAUUAUAAUCCUAG-----AGGCAGCAGGCGGCGACGGGCAAUGCACCUGCAUUGGAUAUUGCGCCUGCAAGUGCAACUUUGCAU (((((((.........)))))))..(((((..((((........((...-----.))..(((((((.(((.(..((((((....))))))..).))))))))))...))))..))))).. ( -43.20) >DroSim_CAF1 18072 115 + 1 UUCUGCACACGCCGCAGGCAGGAGCGCAAAAAUGCAAUUAUAAUCCUAG-----AGGCAGCAGGCGGCGACGGGCAAUGCAACUGCAUUGGAUAUUGCGCCUGCAAGUGCAACUUUGCAU ...(((((..(((.((((...((..(((....)))..)).....))).)-----.))).(((((((.(((.(..((((((....))))))..).))))))))))..)))))......... ( -42.00) >DroEre_CAF1 28489 105 + 1 UUCUGCCCGUGCCCCAGGA-AGAGCGCAAAAAUGCAAUUAUAAUCUCAGGCAGCAGGCGG--------------CAGUGCAACUGCAUUGGAAAUUGCCGCUGCAAGUGCAACUUUGCAU ((((..((........)).-)))).(((((..((((.............(((((.(((((--------------((((((....))))))....))))))))))...))))..))))).. ( -33.29) >DroYak_CAF1 28303 112 + 1 --------AUGCCCCAGGCAGGAGCGCAAAAAUGCAAUUAUAAUCCCAGGCAGCAGGCGGCAGGCGGCGACUGGCAGUGCAACUGCAUUGGAAAUUGCAGCUGGCCGUGCAACUUUGCAU --------..((.((.....)).))(((((..((((.........((........))((((.(((.((((.(..((((((....))))))..).)))).))).))))))))..))))).. ( -37.50) >consensus UUCUGCCCACGCCCCAGGCAGGAGCGCAAAAAUGCAAUUAUAAUCCCAG_____AGGCAGCAGGCGGCGACGGGCAGUGCAACUGCAUUGGAAAUUGCGCCUGCAAGUGCAACUUUGCAU ..........(((....((....))(((....)))....................))).(((((((.(((.(..((((((....))))))..).))))))))))..(((((....))))) (-25.42 = -27.54 + 2.12)

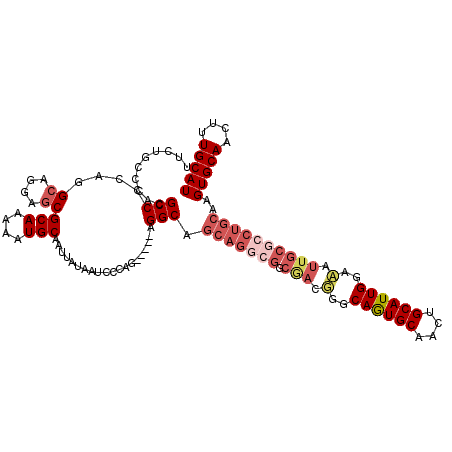

| Location | 5,289,914 – 5,290,029 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.35 |
| Mean single sequence MFE | -41.37 |
| Consensus MFE | -25.26 |
| Energy contribution | -27.22 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849117 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5289914 115 - 22224390 AUGCAAAGUUGCACUUGCAGGCGCAAUUUUCAAUGCAGUUGCACUGCCAGUUGCCGCCUGCUGCCA-----CUGGGAUUAUAAUUGCAUUUUUGCGCUCCUGCCUGGGGCGUGGGCAGAA ..((((((.((((...((((((((((((..((.(((....))).))..))))).)))))))..((.-----...))........)))).))))))((((.((((...)))).)))).... ( -44.50) >DroSec_CAF1 27252 115 - 1 AUGCAAAGUUGCACUUGCAGGCGCAAUAUCCAAUGCAGGUGCAUUGCCCGUCGCCGCCUGCUGCCU-----CUAGGAUUAUAAUUGCAUUUUUGCGCUCCUGCCUGCGGCGUGGGCAGAA .........((((((((((((........))..))))))))))(((((((.((((((..((..((.-----...)).........(((....)))......))..))))))))))))).. ( -45.60) >DroSim_CAF1 18072 115 - 1 AUGCAAAGUUGCACUUGCAGGCGCAAUAUCCAAUGCAGUUGCAUUGCCCGUCGCCGCCUGCUGCCU-----CUAGGAUUAUAAUUGCAUUUUUGCGCUCCUGCCUGCGGCGUGUGCAGAA ..((((((.((((...((((((((.((...((((((....))))))...)).).)))))))..((.-----...))........)))).))))))((.(.((((...)))).).)).... ( -39.70) >DroEre_CAF1 28489 105 - 1 AUGCAAAGUUGCACUUGCAGCGGCAAUUUCCAAUGCAGUUGCACUG--------------CCGCCUGCUGCCUGAGAUUAUAAUUGCAUUUUUGCGCUCU-UCCUGGGGCACGGGCAGAA .(((...(((((....))))).))).........((((.....)))--------------).(((((.(((((.((........((((....))))....-..)).)))))))))).... ( -36.94) >DroYak_CAF1 28303 112 - 1 AUGCAAAGUUGCACGGCCAGCUGCAAUUUCCAAUGCAGUUGCACUGCCAGUCGCCGCCUGCCGCCUGCUGCCUGGGAUUAUAAUUGCAUUUUUGCGCUCCUGCCUGGGGCAU-------- ..((((((.((((.(((((((((((........))))))))....)))((((.((((..((.....)).))..)))))).....)))).))))))(((((.....)))))..-------- ( -40.10) >consensus AUGCAAAGUUGCACUUGCAGGCGCAAUUUCCAAUGCAGUUGCACUGCCAGUCGCCGCCUGCUGCCU_____CUGGGAUUAUAAUUGCAUUUUUGCGCUCCUGCCUGGGGCGUGGGCAGAA ..((((((.((((...(((((((.......((.(((....))).))........)))))))..(((.......)))........)))).))))))(((((.....))))).......... (-25.26 = -27.22 + 1.96)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:46 2006