
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,280,568 – 5,280,705 |
| Length | 137 |
| Max. P | 0.744832 |

| Location | 5,280,568 – 5,280,670 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.11 |
| Mean single sequence MFE | -31.58 |
| Consensus MFE | -18.58 |
| Energy contribution | -18.62 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5280568 102 + 22224390 GCAAACAGUGGCGA---------AGAUGUUGUCUGGACAGCG-GCAGCUGGGAAUGGAU-------GGACAGAAAAGUGCAGCU-UGCAAGGUCAACAUGGCGUAUGCGCGAUGUUUGAG .((((((.(.(((.---------..((((..(...(((....-(((((((.(..((...-------...))......).)))).-)))...)))...)..))))...))).))))))).. ( -31.40) >DroSec_CAF1 17496 99 + 1 GCAAACAGGGGCGA---------AGAUGAUGUCUGGACAGCG-GGAGCGGG--UUGUGU-------GGACAGA-AAGUGCAGUU-UGCAAGGUCAACAUUGCGUAUGCGCGAUGUUUGAG ((((((....(((.---------......((((((.(((((.-.......)--)))).)-------)))))..-...))).)))-)))....(((((((((((....))))))).)))). ( -35.80) >DroSim_CAF1 12251 99 + 1 GCAAACAGGGGCGA---------AGAUGAUGUCUGGACAGCG-GGAGCGGG--UUGUGU-------GGACAGA-AAGUGCAGUU-UGCAAGGUCAACAUUGCGUAUGCGCAAUGUUGGAG ((((((....(((.---------......((((((.(((((.-.......)--)))).)-------)))))..-...))).)))-)))....(((((((((((....))))))))))).. ( -39.00) >DroEre_CAF1 17219 106 + 1 GCAAACAGCGGCGA---------AGAUGAAGACAGGGCAGCGGGCAGCGGGCGCAGGGUGGAA---UGACA-A-AAGUGCAGUUUUGCAAGGCCAACAUUGCGUAUGCGUGAUGUUUGGG .((((((((.((..---------...((....)).....))..)).(((.((((((..(((..---((.((-(-((......)))))))...)))...)))))).)))....)))))).. ( -28.10) >DroYak_CAF1 13614 117 + 1 GCAAACAGGGGCGAAGAAGGUGAAGAUGAAGACAGGACAGCG-GCAGCGGCAGCAUGAUGGAAGCAGGACAAA-AAGUGCAGUU-GGCAAGGUCAACACGGCGUAUGCGCGAUGUUGGAG ((........)).......................((((.((-((((((.(.((((..((.........))..-..)))).(((-(((...))))))..).))).))).)).)))).... ( -23.60) >consensus GCAAACAGGGGCGA_________AGAUGAUGUCUGGACAGCG_GCAGCGGG__CUGGGU_______GGACAGA_AAGUGCAGUU_UGCAAGGUCAACAUUGCGUAUGCGCGAUGUUUGAG ((........)).......................(((.((.....)).............................(((......)))..)))(((((((((....))))))))).... (-18.58 = -18.62 + 0.04)

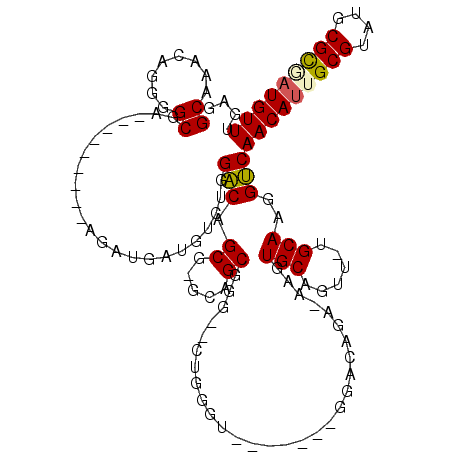
| Location | 5,280,599 – 5,280,705 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.71 |
| Mean single sequence MFE | -30.83 |
| Consensus MFE | -16.58 |
| Energy contribution | -17.14 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5280599 106 - 22224390 UAAUUA-----CCAAGGUGUACAGCGGGUGUUUGAAAGUCCUCAAACAUCGCGCAUACGCCAUGUUGACCUUGCA-AGCUGCACUUUUCUGUCC-------AUCCAUUCCCAGCUGC-CG ...(((-----.((.((((((..((((((((((((......))))))))).))).)))))).)).)))....(((-.((((.............-------.........)))))))-.. ( -29.75) >DroSec_CAF1 17527 106 - 1 UAAUUACAUCACCGAGGUGU--AGCGGGUGUAUUAAAGUCCUCAAACAUCGCGCAUACGCAAUGUUGACCUUGCA-AACUGCACUU-UCUGUCC-------ACACAA--CCCGCUCC-CG ....((((((.....)))))--)((((((((......(((....(((((.(((....))).))))))))..(((.-....)))...-.......-------..)).)--)))))...-.. ( -27.20) >DroSim_CAF1 12282 106 - 1 UAAUUACAUCACCGAGGUGU--AGCGGGUGUAUUAAAGUCCUCCAACAUUGCGCAUACGCAAUGUUGACCUUGCA-AACUGCACUU-UCUGUCC-------ACACAA--CCCGCUCC-CG ....((((((.....)))))--)((((((((............((((((((((....))))))))))....(((.-....)))...-.......-------..)).)--)))))...-.. ( -33.30) >DroEre_CAF1 17250 111 - 1 UAAUUACAGUGCCCAGCUA----GCGGGUGCAUUAAAGUUCCCAAACAUCACGCAUACGCAAUGUUGGCCUUGCAAAACUGCACUU-U-UGUCA---UUCCACCCUGCGCCCGCUGCCCG ...............((.(----(((((((((.........((((.(((..((....))..)))))))...((((....))))...-.-.....---........))))))))))))... ( -30.00) >DroYak_CAF1 13654 117 - 1 UAAUGACAGUGCCCAGGUAGCGAGCGGGUGUAUUAAAGUCCUCCAACAUCGCGCAUACGCCGUGUUGACCUUGCC-AACUGCACUU-UUUGUCCUGCUUCCAUCAUGCUGCCGCUGC-CG ........(.((...(((((((((((((......(((((....((((((.(((....))).)))))).....((.-....))))))-)....))))))).......))))))...))-). ( -33.91) >consensus UAAUUACAGCACCCAGGUGU__AGCGGGUGUAUUAAAGUCCUCAAACAUCGCGCAUACGCAAUGUUGACCUUGCA_AACUGCACUU_UCUGUCC_______ACCCAA__CCCGCUGC_CG ......................((((((.((..........((.(((((.(((....))).)))))))...(((......))).......................)).))))))..... (-16.58 = -17.14 + 0.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:36 2006