
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,279,264 – 5,279,448 |
| Length | 184 |
| Max. P | 0.987580 |

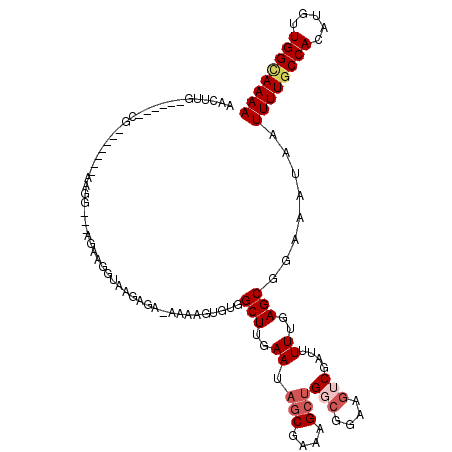
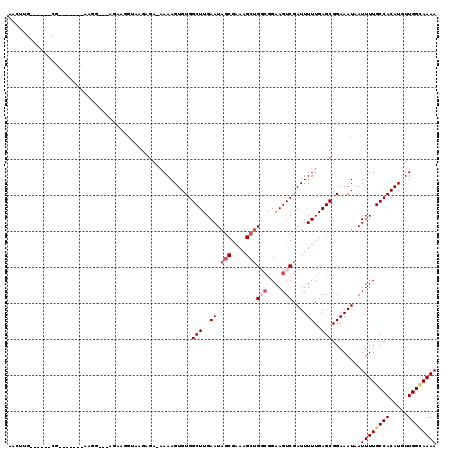
| Location | 5,279,264 – 5,279,368 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.02 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -17.98 |
| Energy contribution | -19.46 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5279264 104 + 22224390 AACUUG------CGAA-----AGGG---CGAAGGUAAGAGC-AAAAGUGUGGCUUGAAUAGCGUAAGCGGGCGGAAGUCGAUUUUUGAGCGGAAAUAAUUUUGCCACAUGUUGGUAAAA ...(((------(...-----...(---(....))....))-))..(((((((..((((.((....)).(((....))).((((((....)))))).)))).))))))).......... ( -27.70) >DroSec_CAF1 16322 101 + 1 AAGAAG------C--------AAGG---AGAAGGUAAGAGA-AAAAGUGUGGCUUGAAUAGCGAAAGCUGGCGGAAGUCGAUUUUUGAGCGGAAAUAAUUUUGCCACAUGUUGGCAAAA .....(------(--------.((.---(...(((((((..-.......(.(((..((.(((....)))(((....)))....))..))).)......)))))))...).)).)).... ( -26.36) >DroSim_CAF1 10976 101 + 1 AAGAAG------C--------AAGG---AGAAGGUAAGGGA-AAAAGUGUGGCUUGAAUAGCGAAAGCUGGCGGAAGUCGAUUUUUGAGCGGAAAUAAUUUUGCCACAUGUUGGCAAAA .....(------(--------.((.---(...(((((((..-.......(.(((..((.(((....)))(((....)))....))..))).)......)))))))...).)).)).... ( -25.56) >DroEre_CAF1 16011 119 + 1 GACUUGGUUUUGCGGAGGGGCAAAGUCAGAAAAAAAUGAGAGAAAAGUGUGGCUUGAAUAGCGAAAGGUGCCGGAAGUCGAUUUUUGAGCGGAAAUAAUUUUGCCACAUGUUGGCAAAA ...(((..(((((......)))))..))).......((...((...(((((((..((((..(....).(((((((((....)))))).)))......)))).))))))).))..))... ( -27.00) >DroYak_CAF1 11905 107 + 1 AACUUG------CGC---AGCAAAG---AAAAAAAAAGAGACGAAAGUGUGGCUUGAAUAGCGAAAGCUGCAGGAAGUCGAUUUUUGAGCGGAAAUAAUUUUGCCACAUGUUGGAAAAA ......------..(---((((...---............((....))(((((..((.((((....))))((((((.....))))))...........))..))))).)))))...... ( -25.50) >consensus AACUUG______CG_______AAGG___AGAAGGUAAGAGA_AAAAGUGUGGCUUGAAUAGCGAAAGCUGGCGGAAGUCGAUUUUUGAGCGGAAAUAAUUUUGCCACAUGUUGGCAAAA ...................................................(((..((.(((....)))(((....)))....))..)))........((((((((.....)))))))) (-17.98 = -19.46 + 1.48)

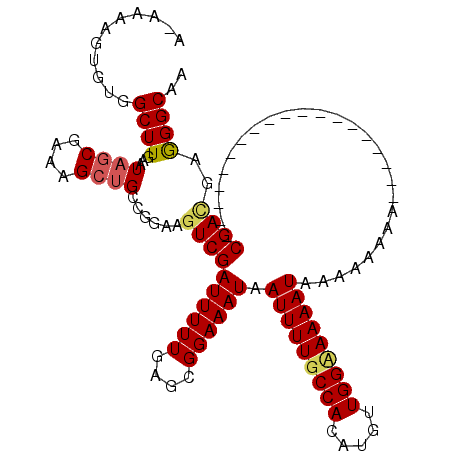
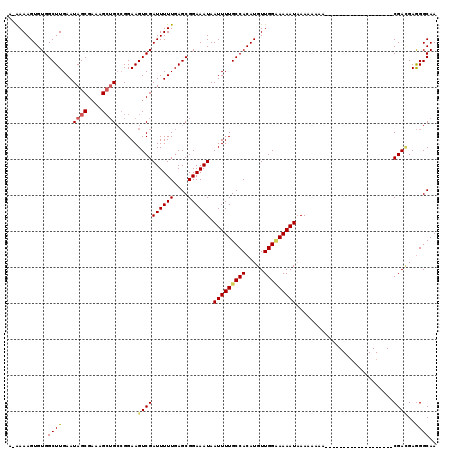
| Location | 5,279,290 – 5,279,408 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -24.75 |
| Consensus MFE | -16.77 |
| Energy contribution | -18.33 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983360 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5279290 118 + 22224390 C-AAAAGUGUGGCUUGAAUAGCGUAAGCGGGCGGAAGUCGAUUUUUGAGCGGAAAUAAUUUUGCCACAUGUUGGUAAAAUCAAAAAAAAAAAGUAAAAAAAUAAACACGAGGAGGGCAA .-....((((.(((..((..((....)).(((....)))....))..))).......(((((((((.....)))))))))........................))))........... ( -27.40) >DroEre_CAF1 16051 101 + 1 AGAAAAGUGUGGCUUGAAUAGCGAAAGGUGCCGGAAGUCGAUUUUUGAGCGGAAAUAAUUUUGCCACAUGUUGGCAAAAUAAAAAACA------------------CCGACGAGGGCAA ...........(((.....)))......((((....((((((((((....)))))).(((((((((.....)))))))))........------------------.))))...)))). ( -24.60) >DroYak_CAF1 11933 100 + 1 ACGAAAGUGUGGCUUGAAUAGCGAAAGCUGCAGGAAGUCGAUUUUUGAGCGGAAAUAAUUUUGCCACAUGUUGGAAAAAUAAAAAAAA-------------------CGAUGAAGGCAA .(((((((.((((((...((((....))))....))))))))))))).............(((((.((((((..............))-------------------).)))..))))) ( -22.24) >consensus A_AAAAGUGUGGCUUGAAUAGCGAAAGCUGCCGGAAGUCGAUUUUUGAGCGGAAAUAAUUUUGCCACAUGUUGGAAAAAUAAAAAAAA___________________CGACGAGGGCAA ...........((((...((((....))))......((((((((((....)))))).(((((((((.....)))))))))...........................))))..)))).. (-16.77 = -18.33 + 1.56)

| Location | 5,279,329 – 5,279,448 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -20.21 |
| Consensus MFE | -16.62 |
| Energy contribution | -16.84 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5279329 119 + 22224390 AUUUUUGAGCGGAAAUAAUUUUGCCACAUGUUGGUAAAAUCAAAAAAAAAAAGUAAAAAAAUAAACACGAGGAGGGCAAUUCUAAAACAAAGUCACGCGCAGAAAAUCAGGGUGAAAGA .((((((.(((((....(((((((((.....))))))))).............................((((......)))).........)).))).)))))).............. ( -18.50) >DroEre_CAF1 16091 101 + 1 AUUUUUGAGCGGAAAUAAUUUUGCCACAUGUUGGCAAAAUAAAAAACA------------------CCGACGAGGGCAAUUCUAAAACAAAGUCACGCGCAGAAAAUCUGAGCGAAAGA .((((((.(((......(((((((((.....)))))))))........------------------..(((...(............)...))).))).)))))).......(....). ( -22.70) >DroYak_CAF1 11973 100 + 1 AUUUUUGAGCGGAAAUAAUUUUGCCACAUGUUGGAAAAAUAAAAAAAA-------------------CGAUGAAGGCAAUACUAAAACAAAGUCACGCUCAGAAAAUGGGGCUGAAAGG .((((((((((.........(((((.((((((..............))-------------------).)))..))))).(((.......)))..)))))))))).............. ( -19.44) >consensus AUUUUUGAGCGGAAAUAAUUUUGCCACAUGUUGGAAAAAUAAAAAAAA___________________CGACGAGGGCAAUUCUAAAACAAAGUCACGCGCAGAAAAUCAGGGUGAAAGA .((((((.(((......(((((((((.....)))))))))..................................(((..............))).))).)))))).............. (-16.62 = -16.84 + 0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:34 2006