
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,278,457 – 5,278,615 |
| Length | 158 |
| Max. P | 0.999165 |

| Location | 5,278,457 – 5,278,575 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.36 |
| Mean single sequence MFE | -37.58 |
| Consensus MFE | -23.27 |
| Energy contribution | -23.67 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5278457 118 + 22224390 GCUAAUGCUUAAUCCGCUCUUAAGCGCUUUUAAGAGAAAUACA-AAUACUGGGAACAGCAGUGGGAACACUAAGCGGCUGCGAACAC-UGUUCCCAGUACCCCAGGCAACUGUGCCCGAU ((....))........((((((((....)))))))).......-..((((((((((((.((((....))))..((....)).....)-))))))))))).....((((....)))).... ( -38.30) >DroSec_CAF1 15517 107 + 1 GCUAAUGUUUAAGCCGCUCUUAAGCGCUUUUAAGAGAG--CCA-AAUACUGGGAACAGCAGUGGGAACACUAUGCGACUGGGAAC----------AGUACCCCAGGCAACUGUGCCCGAU (((.......((((.(((....))))))).......))--)..-.....((((.((((.((((....)))).(((..(((((...----------.....)))))))).)))).)))).. ( -34.14) >DroSim_CAF1 10171 107 + 1 GCUAAUGUUUAAGCCGCUCUUAAGCGCUUUUAAGAGAG--CCA-AAUACUGGGAACAGCAGUGGGAACACUGCGCGACUGUGAAC----------AGUACCCCAGGCAACUGUGCCCGAU (((.......((((.(((....))))))).......))--)..-....(((((....((((((....))))))...((((....)----------)))..)))))(((....)))..... ( -36.24) >DroEre_CAF1 15105 112 + 1 ---AAAGUUUUCCGCGCUCUUAGGCGCUUUUAAGAGAG--CCA-AAUACUGGGAACAGCAGUGGGAACACCAUGCGGGUGGGAACACCU--UUGCAGUGCCCCAAGCAACUGUCCCCGAA ---..(((.((..((.((((((((....)))))))).)--)..-)).)))(((.((((...((((..(((..((((((((....)))))--..)))))).)))).....)))).)))... ( -38.70) >DroYak_CAF1 11052 115 + 1 ---AAAUUUUUUCCUGCUCUUAGGGCAUUUUAAGAGAG--CCAAAAUACUGGGAACAGUAGUGGGAACACCAUGCGGGUGGGAACACAUGCUUGCAGUACCCCAAGCCACUGUUCCCACA ---.........((..((((((((....)))))))).(--(.....(((((....)))))((((.....))))))))(((((((((...(((((........)))))...))))))))). ( -40.50) >consensus GCUAAUGUUUAACCCGCUCUUAAGCGCUUUUAAGAGAG__CCA_AAUACUGGGAACAGCAGUGGGAACACUAUGCGGCUGGGAACAC_U__U__CAGUACCCCAGGCAACUGUGCCCGAU ................((((((((....)))))))).............((((.((((..(((....)))...((..(((((..................)))))))..)))).)))).. (-23.27 = -23.67 + 0.40)



| Location | 5,278,457 – 5,278,575 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.36 |
| Mean single sequence MFE | -39.96 |
| Consensus MFE | -23.24 |
| Energy contribution | -24.04 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5278457 118 - 22224390 AUCGGGCACAGUUGCCUGGGGUACUGGGAACA-GUGUUCGCAGCCGCUUAGUGUUCCCACUGCUGUUCCCAGUAUU-UGUAUUUCUCUUAAAAGCGCUUAAGAGCGGAUUAAGCAUUAGC ..((((((....))))))((((((((((((((-((....((....))..((((....)))))))))))))))))))-).................((((((.......))))))...... ( -42.40) >DroSec_CAF1 15517 107 - 1 AUCGGGCACAGUUGCCUGGGGUACU----------GUUCCCAGUCGCAUAGUGUUCCCACUGCUGUUCCCAGUAUU-UGG--CUCUCUUAAAAGCGCUUAAGAGCGGCUUAAACAUUAGC ...(((.((((((((((((((....----------..))))))..))).((((....))))))))).))).((...-.((--(((((((((......))))))).))))...))...... ( -35.60) >DroSim_CAF1 10171 107 - 1 AUCGGGCACAGUUGCCUGGGGUACU----------GUUCACAGUCGCGCAGUGUUCCCACUGCUGUUCCCAGUAUU-UGG--CUCUCUUAAAAGCGCUUAAGAGCGGCUUAAACAUUAGC ...(((.(((((.....((..((((----------((.(......).))))))..))....))))).))).((...-.((--(((((((((......))))))).))))...))...... ( -35.60) >DroEre_CAF1 15105 112 - 1 UUCGGGGACAGUUGCUUGGGGCACUGCAA--AGGUGUUCCCACCCGCAUGGUGUUCCCACUGCUGUUCCCAGUAUU-UGG--CUCUCUUAAAAGCGCCUAAGAGCGCGGAAAACUUU--- ...(((((((((....(((((((((((..--.((((....)))).))..)))).)))))..)))))))))(((.((-(.(--(.((((((........)))))).)).))).)))..--- ( -45.10) >DroYak_CAF1 11052 115 - 1 UGUGGGAACAGUGGCUUGGGGUACUGCAAGCAUGUGUUCCCACCCGCAUGGUGUUCCCACUACUGUUCCCAGUAUUUUGG--CUCUCUUAAAAUGCCCUAAGAGCAGGAAAAAAUUU--- ..((((((((((((..(((((....(((..((((((........)))))).))))))))))))))))))))...((((..--((((((((........)))))).))..))))....--- ( -41.10) >consensus AUCGGGCACAGUUGCCUGGGGUACUG__A__A_GUGUUCCCAGCCGCAUAGUGUUCCCACUGCUGUUCCCAGUAUU_UGG__CUCUCUUAAAAGCGCUUAAGAGCGGCUUAAACAUUAGC ...(((.(((((.((((((((.((...........))))))))..))..((((....))))))))).)))..............(((((((......)))))))................ (-23.24 = -24.04 + 0.80)

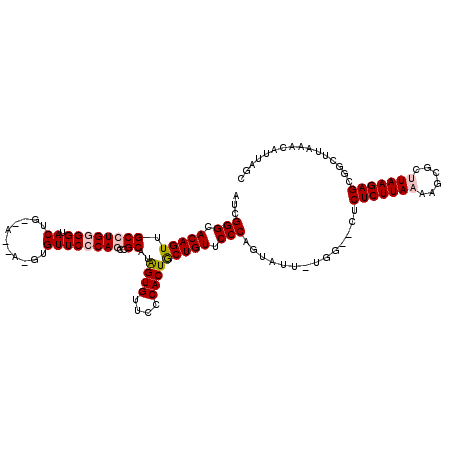

| Location | 5,278,497 – 5,278,615 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.60 |
| Mean single sequence MFE | -38.73 |
| Consensus MFE | -18.81 |
| Energy contribution | -19.60 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5278497 118 + 22224390 ACA-AAUACUGGGAACAGCAGUGGGAACACUAAGCGGCUGCGAACAC-UGUUCCCAGUACCCCAGGCAACUGUGCCCGAUUUCUAAUUACUGUGUUGUUGGUUGGUUGGUGGGUGGUGGG ...-..((((((((((((.((((....))))..((....)).....)-)))))))))))((((((....))).(((((....((((((((......).)))))))....)))))...))) ( -45.90) >DroEre_CAF1 15140 96 + 1 CCA-AAUACUGGGAACAGCAGUGGGAACACCAUGCGGGUGGGAACACCU--UUGCAGUGCCCCAAGCAACUGUCCCCGAACUCCAAUUACUGGGUAGGC--------------------- ((.-.....((((.((((...((((..(((..((((((((....)))))--..)))))).)))).....)))).)))).((.(((.....))))).)).--------------------- ( -31.90) >DroYak_CAF1 11087 99 + 1 CCAAAAUACUGGGAACAGUAGUGGGAACACCAUGCGGGUGGGAACACAUGCUUGCAGUACCCCAAGCCACUGUUCCCACAUUCUAAUUACUGGGUGGGC--------------------- (((...(((((....))))).))).....((((.((((((((((((...(((((........)))))...)))))))))..........))).))))..--------------------- ( -38.40) >consensus CCA_AAUACUGGGAACAGCAGUGGGAACACCAUGCGGGUGGGAACAC_UG_UUGCAGUACCCCAAGCAACUGUCCCCGAAUUCUAAUUACUGGGUAGGC_____________________ ((.......((((.((((...((((.((.....(((.(((....))).))).....))..)))).....)))).))))(((((........)))))))...................... (-18.81 = -19.60 + 0.79)


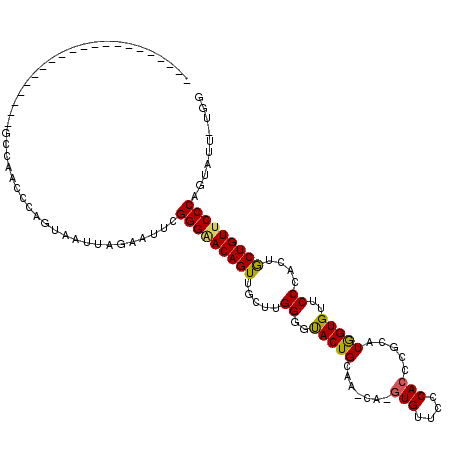
| Location | 5,278,497 – 5,278,615 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.60 |
| Mean single sequence MFE | -36.40 |
| Consensus MFE | -23.64 |
| Energy contribution | -23.53 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.65 |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.999165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5278497 118 - 22224390 CCCACCACCCACCAACCAACCAACAACACAGUAAUUAGAAAUCGGGCACAGUUGCCUGGGGUACUGGGAACA-GUGUUCGCAGCCGCUUAGUGUUCCCACUGCUGUUCCCAGUAUU-UGU ......((((............((......))..........((((((....))))))))))((((((((((-((....((....))..((((....))))))))))))))))...-... ( -37.90) >DroEre_CAF1 15140 96 - 1 ---------------------GCCUACCCAGUAAUUGGAGUUCGGGGACAGUUGCUUGGGGCACUGCAA--AGGUGUUCCCACCCGCAUGGUGUUCCCACUGCUGUUCCCAGUAUU-UGG ---------------------.((.((((((...)))).))..(((((((((....(((((((((((..--.((((....)))).))..)))).)))))..)))))))))......-.)) ( -33.70) >DroYak_CAF1 11087 99 - 1 ---------------------GCCCACCCAGUAAUUAGAAUGUGGGAACAGUGGCUUGGGGUACUGCAAGCAUGUGUUCCCACCCGCAUGGUGUUCCCACUACUGUUCCCAGUAUUUUGG ---------------------..............(((((((((((((((((((..(((((....(((..((((((........)))))).)))))))))))))))))))).))))))). ( -37.60) >consensus _____________________GCCAACCCAGUAAUUAGAAUUCGGGAACAGUUGCUUGGGGUACUGCAA_CA_GUGUUCCCACCCGCAUGGUGUUCCCACUGCUGUUCCCAGUAUU_UGG ...........................................(((((((((.....((..(((((.......(((....))).....)))))..))....))))))))).......... (-23.64 = -23.53 + -0.10)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:32 2006