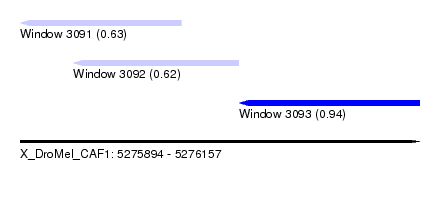
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,275,894 – 5,276,157 |
| Length | 263 |
| Max. P | 0.935421 |

| Location | 5,275,894 – 5,276,000 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 85.78 |
| Mean single sequence MFE | -33.30 |
| Consensus MFE | -24.19 |
| Energy contribution | -26.12 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5275894 106 - 22224390 AUCGCAGUCGAUCAGCUCUUGGGAAACACGCUCCCGAUCCUGAUCCUGCUGCUGCUGCUGCUGCAGUCUGGACAACUCGCUCAUGUUGAUA-----GUAUCUUCAAUGCGA .((((.(((((((((...((((((.......))))))..))))))..(((((.((....)).)))))...)))...........(((((..-----......))))))))) ( -35.20) >DroSec_CAF1 12796 111 - 1 AUCGCAGUCGAUCAGCUCUUGGGAAACACGCUCCCGAUCCUGAUCCUGCUGCUGCUGCUGCUGCAGUCUGGACAACUCGCUCAUGUUGAUAUGUAUGUAUCUUCAAUGCGA .((((((((((((((...((((((.......))))))..))))))..(((((.((....)).)))))...)))..........((..(((((....)))))..)).))))) ( -36.90) >DroSim_CAF1 7461 111 - 1 AUCGCAGUCGAUCAGCUCUUGGGAAACACGCUCCCGAUCCUGAUCCUGCUGCUGCUGCUGCUGCAGUCUGGACAACUCGCUCAUGUUGAUAUGUAAGUAUCUUCUAUGCGA .((((((((((((((...((((((.......))))))..))))))..(((((.((....)).)))))...)))..............(((((....))))).....))))) ( -36.10) >DroYak_CAF1 8284 91 - 1 AUCGCAGUCGAUCAGCUCCUGGGGAACUUGCUCCUGAUCCU------------GCUGCUGCUGCAGUCUGGACAACUCGCUCAUGUUGAUC--------UCUUCUUUGGGA .((.(((..(((((((....((((......))))((.(((.------------(((((....)))))..)))))..........)))))))--------......))).)) ( -25.00) >consensus AUCGCAGUCGAUCAGCUCUUGGGAAACACGCUCCCGAUCCUGAUCCUGCUGCUGCUGCUGCUGCAGUCUGGACAACUCGCUCAUGUUGAUA_____GUAUCUUCAAUGCGA .((((((((((((((...((((((.......))))))..))))))........(((((....)))))...)))..............(((((....))))).....))))) (-24.19 = -26.12 + 1.94)

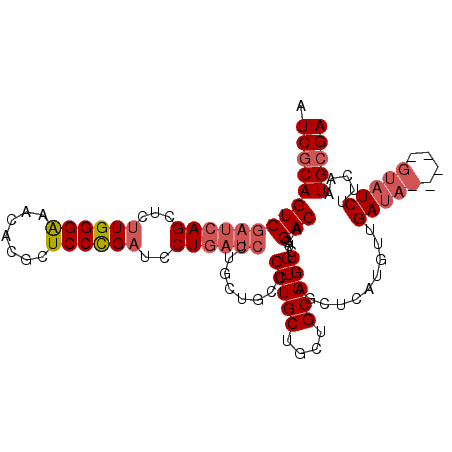
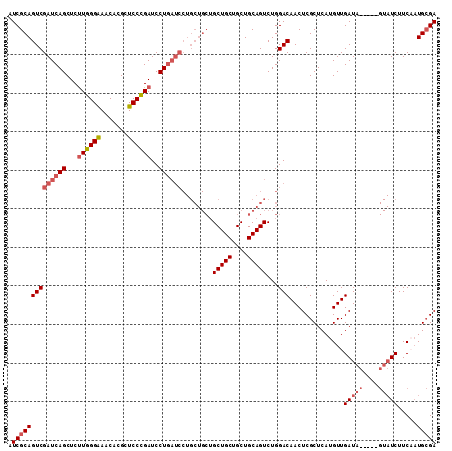
| Location | 5,275,929 – 5,276,038 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 85.15 |
| Mean single sequence MFE | -34.40 |
| Consensus MFE | -26.83 |
| Energy contribution | -29.20 |
| Covariance contribution | 2.37 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5275929 109 - 22224390 --UCCUCCACCUUGGAGGUCGUAUCCUCCACAAAGUCAAUAUCGCAGUCGAUCAGCUCUUGGGAAACACGCUCCCGAUCCUGAUCCUGCUGCUGCUGCUGCUGCAGUCUGG --....(((...((((((......)))))).............(((((.((((((...((((((.......))))))..))))))..))))).(((((....))))).))) ( -38.40) >DroSec_CAF1 12836 109 - 1 --UCCCCCACCUUGGAGGCCGCCACCUCCUCAAAGUCAAUAUCGCAGUCGAUCAGCUCUUGGGAAACACGCUCCCGAUCCUGAUCCUGCUGCUGCUGCUGCUGCAGUCUGG --....(((....(((((......)))))..............(((((.((((((...((((((.......))))))..))))))..))))).(((((....))))).))) ( -37.10) >DroSim_CAF1 7501 109 - 1 --UCCCCCACCUUGGAGGCCGCCACCUCCUCAAAGUCAAUAUCGCAGUCGAUCAGCUCUUGGGAAACACGCUCCCGAUCCUGAUCCUGCUGCUGCUGCUGCUGCAGUCUGG --....(((....(((((......)))))..............(((((.((((((...((((((.......))))))..))))))..))))).(((((....))))).))) ( -37.10) >DroYak_CAF1 8316 99 - 1 CAACCCCCACCUCGGAGGUCGUAUCAUCCAAAGAGGCAAUAUCGCAGUCGAUCAGCUCCUGGGGAACUUGCUCCUGAUCCU------------GCUGCUGCUGCAGUCUGG ..(((.((.....)).))).(((...(((..((..((...((((....))))..))..))..)))...))).......((.------------(((((....)))))..)) ( -25.00) >consensus __UCCCCCACCUUGGAGGCCGCAACCUCCACAAAGUCAAUAUCGCAGUCGAUCAGCUCUUGGGAAACACGCUCCCGAUCCUGAUCCUGCUGCUGCUGCUGCUGCAGUCUGG ......(((....(((((......)))))..............(((((.((((((...((((((.......))))))..))))))..))))).(((((....))))).))) (-26.83 = -29.20 + 2.37)

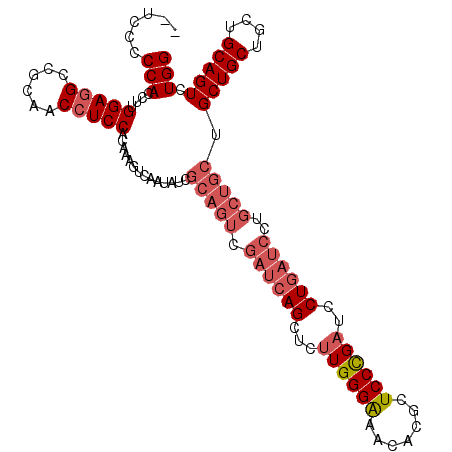
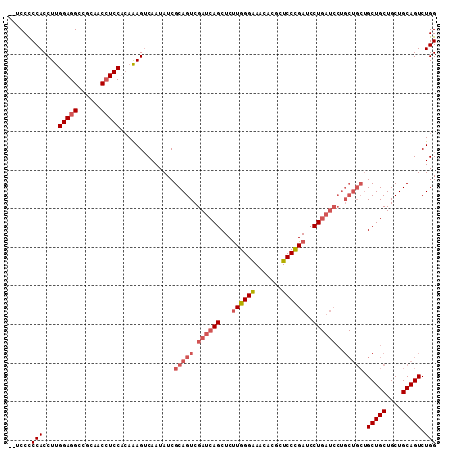
| Location | 5,276,038 – 5,276,157 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.84 |
| Mean single sequence MFE | -44.70 |
| Consensus MFE | -37.48 |
| Energy contribution | -38.40 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
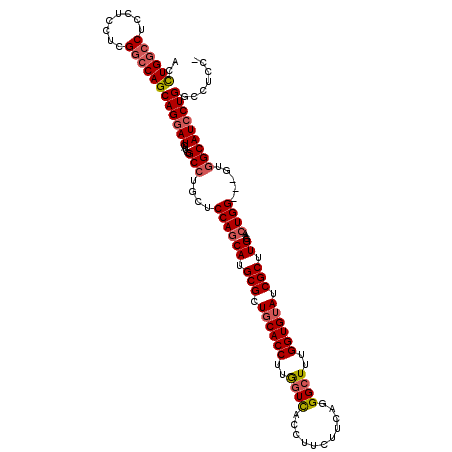
>X_DroMel_CAF1 5276038 119 - 22224390 ACCUGGCCUCCUCCUCGGCCAGCAGGAUCAUGGCCUGCUCCAGCAUGCGCUGCACCUUGGUCACCUUCUUCAGGGCUUUGGUGUAUCGCUUGGAACUGGCUGGUGGCAUCCUGGCCUCC- ....((((....((((((((((((((.......)))))(((((...(((.((((((..(((..(((.....))))))..)))))).))))))))..))))))).))......))))...- ( -49.10) >DroSec_CAF1 12945 116 - 1 ACCUGGCCUCUUCCUCGGCCAGCAGGAUCAUGGCCUGCUCCAGCAUGCGCUGCACCUUGGUCACCAUCUUCAGGGCUUUGGUGUAUCGCUUGGAACUGG---GUGGCAUCCUGGCCUCC- ..((((((........))))))((((((....(((.((.((((((.(((.((((((..((((.(........)))))..)))))).))).))...))))---)))))))))))......- ( -47.20) >DroSim_CAF1 7610 116 - 1 ACCUGGCCUCUUCCUCGGCCAGCAGGAUCAUGGCCUGCUCCAGCAUGCGCUGCACCUUGGUCACCUUCUUCAGGGCUUUGGUGUAUCGCUUGGAACUGG---GUGGCAUCCUGGCCUCC- ..((((((........))))))((((((....(((.((.((((((.(((.((((((..(((..(((.....))))))..)))))).))).))...))))---)))))))))))......- ( -47.40) >DroEre_CAF1 12732 112 - 1 ACCUGGCCUCCUCCUCGGCCAGCAGGAUCAGGGCCUGGUCCAGCAUGCGCUGCACCUUGCUUACCUUCUGCAGGGCUUUGGUGUAUCGCUUGGAAGUGG---GUGGCAUCCUGAC----- ..((((((........))))))(((((((...(((((.((((((((((((((..((((((.........))))))...)))))))).)).))))..)))---)).).))))))..----- ( -46.80) >DroYak_CAF1 8535 117 - 1 ACUUGAACUCCUCCUCGGACAGCAGAAUCAGUGCGUGUCCCAGCAUGCGCUCCACCUUAGUCACCUUAUUCAUGGCUUUGGUGUAUCGCUUGUUACUGG---AUGGCAUCCUGACAUCGC .........(((((...((((((......(((((((((....))))))))).((((..(((((.........)))))..))))....)).))))...))---).)).............. ( -33.00) >consensus ACCUGGCCUCCUCCUCGGCCAGCAGGAUCAUGGCCUGCUCCAGCAUGCGCUGCACCUUGGUCACCUUCUUCAGGGCUUUGGUGUAUCGCUUGGAACUGG___GUGGCAUCCUGGCCUCC_ ..((((((........))))))((((((....(((....((((((.(((.((((((..((((...........))))..)))))).))).))...)))).....)))))))))....... (-37.48 = -38.40 + 0.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:27 2006