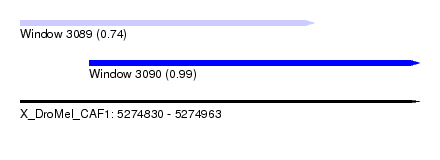
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,274,830 – 5,274,963 |
| Length | 133 |
| Max. P | 0.992095 |

| Location | 5,274,830 – 5,274,928 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.96 |
| Mean single sequence MFE | -24.38 |
| Consensus MFE | -11.84 |
| Energy contribution | -12.16 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5274830 98 + 22224390 GCAUUGGAAUUUAAGUUCCAA-----------------UAUGAAA--AAUGUCAUUUACAGA---UGGAUUAUCUACUUAGUGCAUGAGUGAUAACUAAUCAUGGAGCAUUAUGAGCAUA ((((((((((....)))))))-----------------)......--(((((......((((---(((.(((((.(((((.....)))))))))))).))).))..)))))....))... ( -20.90) >DroSec_CAF1 11720 103 + 1 GCAUUGGAAUUUAAGUUCCAA-----------------UAUGAUGUGAUGAUCAGUUACAGAUGAUGGAUGAUCUACUUAGUGCAUGAGUGAUAACUGAUCAUGGAGCAUUAUGGGGAUG ..((((((((....)))))))-----------------)(((((((.(((((((((((.((((........))))(((((.....)))))..)))))))))))...)))))))....... ( -31.20) >DroSim_CAF1 5699 120 + 1 GCAUUGGAAUCUAAGUUCUAUCUAAGAUGUGAUGAAAUGAUGAUAUGAUGAUCAUUUACAGAUGAUGGAUGAUCUACUUAGUGCAUGAGUGAUAACUGAUCAUGGAGCAUUAUGGGGAUG .(((((..((((.((......)).)))).)))))..((((((.((((((((((((((((....).)))))))))((((((.....)))))).......))))))...))))))....... ( -26.10) >DroEre_CAF1 11567 99 + 1 AAAAUUGAAUUUAAAUGCAGG-C----------CAG--UAU--AA--AAUGUCAUUUACAGAUG----AUGAUCUUCCUAGUGCACGAGUGAUAACUGAUCAUGGAGCAUUAUGAGGAUC ..............((((...-.----------..)--)))--..--...((((((....))))----))(((((((.((((((.(..(((((.....))))).).)))))).))))))) ( -23.60) >DroYak_CAF1 7207 108 + 1 AAAUUGGAAUUUAAAUGCAGG-UACGAUAUGGUCAG---AA--AA--AAUGUCAUUUACAGAUG----AUGAUUUACUUAGUUCACUAGUGAUAACUGAUCAUGGAGCAUUACGAGGAUC .............(((((...-.....(((((((((---..--..--...((((((....))))----))...(((((.((....)))))))...)))))))))..)))))......... ( -20.10) >consensus GCAUUGGAAUUUAAGUUCAAA_____________A___UAUGAAA__AAUGUCAUUUACAGAUG_UGGAUGAUCUACUUAGUGCAUGAGUGAUAACUGAUCAUGGAGCAUUAUGAGGAUC ....((((((....))))))..................................................(((((.(.((((((....(((((.....)))))...)))))).).))))) (-11.84 = -12.16 + 0.32)


| Location | 5,274,853 – 5,274,963 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.48 |
| Mean single sequence MFE | -29.96 |
| Consensus MFE | -15.76 |
| Energy contribution | -16.52 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5274853 110 + 22224390 UGAAA--AAUGUCAUUUACAGA---UGGAUUAUCUACUUAGUGCAUGAGUGAUAACUAAUCAUGGAGCAUUAUGAGCAUAUCCAUCUGAGAAUGUAACGAAUGAUCCCUGAUCCC----- .....--..((((((((.((((---(((((...((...((((((....(((((.....)))))...))))))..))...))))))))).)))))..)))...((((...))))..----- ( -29.40) >DroSec_CAF1 11743 108 + 1 UGAUGUGAUGAUCAGUUACAGAUGAUGGAUGAUCUACUUAGUGCAUGAGUGAUAACUGAUCAUGGAGCAUUAUGGGGAUGUCCAUCUGACAAUGUAACGAAUGAUCCC------------ ......(((.((..((((((...(((((((.((((...((((((....(((((.....)))))...))))))...)))))))))))......))))))..)).)))..------------ ( -33.50) >DroSim_CAF1 5739 108 + 1 UGAUAUGAUGAUCAUUUACAGAUGAUGGAUGAUCUACUUAGUGCAUGAGUGAUAACUGAUCAUGGAGCAUUAUGGGGAUGUCCAUUUGACAAUGUAUCGAAUUAUCCC------------ ......((((((....((((...(((((((.((((...((((((....(((((.....)))))...))))))...)))))))))))......))))....))))))..------------ ( -28.10) >DroEre_CAF1 11594 99 + 1 U--AA--AAUGUCAUUUACAGAUG----AUGAUCUUCCUAGUGCACGAGUGAUAACUGAUCAUGGAGCAUUAUGAGGAUCUCCAUCUGAGUCUGAUCCGAAUGAUCC------------- .--..--...(((((((.((((((----..(((((((.((((((.(..(((((.....))))).).)))))).)))))))..)))))).(......).)))))))..------------- ( -31.80) >DroYak_CAF1 7243 112 + 1 A--AA--AAUGUCAUUUACAGAUG----AUGAUUUACUUAGUUCACUAGUGAUAACUGAUCAUGGAGCAUUACGAGGAUCUUCAUCUGAGUCUGUAGCGAAUGCUCCCAGAUCUCUGAUC .--..--...........((((..----.....(((((.((....))))))).....((((..((((((((.((.((((..((....))))))....))))))))))..))))))))... ( -27.00) >consensus UGAAA__AAUGUCAUUUACAGAUG_UGGAUGAUCUACUUAGUGCAUGAGUGAUAACUGAUCAUGGAGCAUUAUGAGGAUCUCCAUCUGAGAAUGUAACGAAUGAUCCC____________ ..................((((((......(((((.(.((((((....(((((.....)))))...)))))).).)))))..))))))................................ (-15.76 = -16.52 + 0.76)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:25 2006