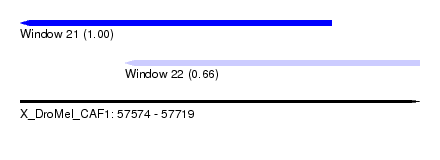
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 57,574 – 57,719 |
| Length | 145 |
| Max. P | 0.997952 |

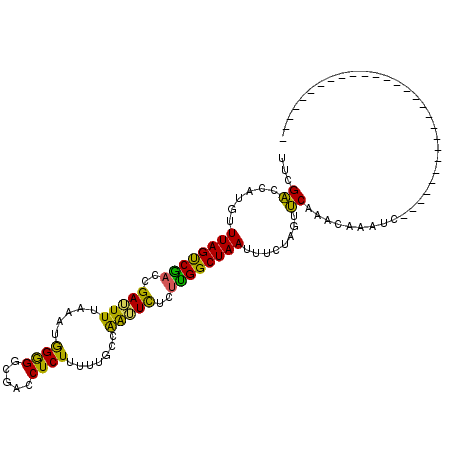
| Location | 57,574 – 57,687 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 66.24 |
| Mean single sequence MFE | -20.67 |
| Consensus MFE | -14.87 |
| Energy contribution | -13.11 |
| Covariance contribution | -1.76 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 57574 113 - 22224390 UUCGGACAUCUUUAGCCGAAUGAUUUUCAAUAGAGACGACCUCUUUUUAGCAAAUUUCUCGGCUAAUUUCUAGUCCAAAAAAAUCUGUUUUGGAACAUCAUUGAUGAAGGUAA ......((((.((((((((..(((((.(...((((.....)))).....).)))))..)))))))).......(((((((.......)))))))........))))....... ( -25.80) >DroEre_CAF1 17122 85 - 1 UUCGACCAUGUUUAGUCGACCGAAUUUUAAUGGGGGCGACCUCUUUUUGCCGGUUCUUUUGGCUAAUUUCUAGUUCAAACAAAUC---------------------------- .(((((........)))))..(((((..(((((((....)))).....(((((.....)))))..)))...))))).........---------------------------- ( -19.60) >DroYak_CAF1 28206 85 - 1 UUCGACCAUGUUUAGUCACCCGAUUUCAAAUGGGGGCGACCUCUUUUUGCCAAUUCUCUUGGCUAAUUUCUAGUUCAAACAAAUC---------------------------- ........(((((.(((.(((.((.....)).)))..)))........(((((.....))))).............)))))....---------------------------- ( -16.60) >consensus UUCGACCAUGUUUAGUCGACCGAUUUUAAAUGGGGGCGACCUCUUUUUGCCAAUUCUCUUGGCUAAUUUCUAGUUCAAACAAAUC____________________________ ...((......((((((((..(((((.....((((.....)))).......)))))..))))))))........))..................................... (-14.87 = -13.11 + -1.76)

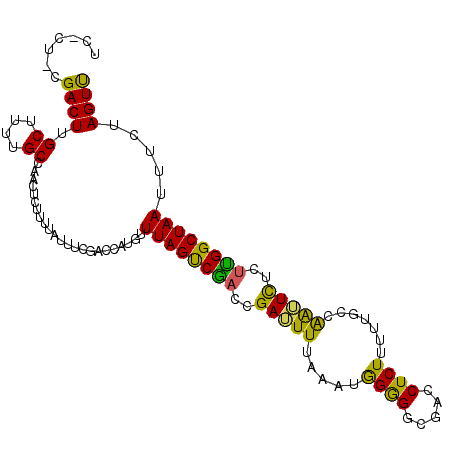
| Location | 57,612 – 57,719 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 79.75 |
| Mean single sequence MFE | -21.10 |
| Consensus MFE | -16.30 |
| Energy contribution | -14.77 |
| Covariance contribution | -1.54 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 57612 107 - 22224390 UGACUCCGACUUGCUUUUGCCAACUUAUUUUUUUCGGACAUCUUUAGCCGAAUGAUUUUCAAUAGAGACGACCUCUUUUUAGCAAAUUUCUCGGCUAAUUUCUAGUC .((((((((...((....)).............))))).....((((((((..(((((.(...((((.....)))).....).)))))..))))))))......))) ( -21.69) >DroEre_CAF1 17132 107 - 1 UCGCUUCGACUUGCUUUUGCCAACUCUUUUAUUUCGACCAUGUUUAGUCGACCGAAUUUUAAUGGGGGCGACCUCUUUUUGCCGGUUCUUUUGGCUAAUUUCUAGUU .......((((.......(((((..........(((((........)))))(((.........((((....)))).......))).....)))))........)))) ( -21.65) >DroYak_CAF1 28216 103 - 1 UC----CGACUUGCUUUUGCCAACUCUUUUAUUUCGACCAUGUUUAGUCACCCGAUUUCAAAUGGGGGCGACCUCUUUUUGCCAAUUCUCUUGGCUAAUUUCUAGUU ..----.((((.......(((((..............((((....((((....))))....)))).(((((.......))))).......)))))........)))) ( -19.96) >consensus UC_CU_CGACUUGCUUUUGCCAACUCUUUUAUUUCGACCAUGUUUAGUCGACCGAUUUUAAAUGGGGGCGACCUCUUUUUGCCAAUUCUCUUGGCUAAUUUCUAGUU .......((((.((....)).......................((((((((..(((((.....((((.....)))).......)))))..)))))))).....)))) (-16.30 = -14.77 + -1.54)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:27 2006