
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,266,048 – 5,266,204 |
| Length | 156 |
| Max. P | 0.903026 |

| Location | 5,266,048 – 5,266,165 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.79 |
| Mean single sequence MFE | -34.95 |
| Consensus MFE | -29.73 |
| Energy contribution | -29.52 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.85 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5266048 117 + 22224390 UUUCUAGAAAGCCGAAAAACGUGUGCACGGAGUGCUGGUUAUCAAAUCGAAUGGCAGCACAUGCACAAACAGGGCUGCCCACGCUCAUCGAUAGGUAUCGAUUGGGGCA---AUAGGUAU ..((((...((((........((((((....((((((.(..((.....))..).)))))).)))))).....))))((((.((...((((((....)))))))))))).---.))))... ( -35.52) >DroSim_CAF1 21365 120 + 1 UUUCUAGAAAGCCGAAAAACGUGUGCUCGAAGUGCUGGUCUUCUAAUCGAAUGGCAGCACAUGUACAAACAGGGCUGCCCACGUUCAUCGAUAUGUAUCGAUAUGGGCAUAAGUACAUAU ..........(((....((((((.((((...((((((.(.(((.....))).).)))))).(((....)))))))....)))))).((((((....))))))...)))............ ( -33.90) >DroYak_CAF1 22413 117 + 1 UUUGUACAGAGCCAAAAAACGUGUGUACCGAGUGCUGGUCAUCGAAUCGAAUGGCAGCACAUGCACAGACAGGGCUGCUCACGCUUGUCGAUAGGUAUCGAUUGAGGCC---GGGCAACU .........((((........((((((....((((((.((((((...)).)))))))))).)))))).....))))((((..((((((((((....))))))..)))).---)))).... ( -35.42) >consensus UUUCUAGAAAGCCGAAAAACGUGUGCACGGAGUGCUGGUCAUCAAAUCGAAUGGCAGCACAUGCACAAACAGGGCUGCCCACGCUCAUCGAUAGGUAUCGAUUGGGGCA___GUACAUAU .........((((........((((((....((((((.(..((.....))..).)))))).)))))).....))))((((......((((((....))))))..))))............ (-29.73 = -29.52 + -0.21)



| Location | 5,266,048 – 5,266,165 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.79 |
| Mean single sequence MFE | -35.13 |
| Consensus MFE | -29.86 |
| Energy contribution | -29.87 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5266048 117 - 22224390 AUACCUAU---UGCCCCAAUCGAUACCUAUCGAUGAGCGUGGGCAGCCCUGUUUGUGCAUGUGCUGCCAUUCGAUUUGAUAACCAGCACUCCGUGCACACGUUUUUCGGCUUUCUAGAAA ....(((.---.(((...((((((....))))))((((((((((((((.(((....))).).)))))).................((((...)))).)))))))...)))....)))... ( -36.00) >DroSim_CAF1 21365 120 - 1 AUAUGUACUUAUGCCCAUAUCGAUACAUAUCGAUGAACGUGGGCAGCCCUGUUUGUACAUGUGCUGCCAUUCGAUUAGAAGACCAGCACUUCGAGCACACGUUUUUCGGCUUUCUAGAAA ...........(((((((((((((....))))))....)))))))(((.((((((.....((((((...(((.....)))...))))))..))))))...(.....)))).......... ( -34.30) >DroYak_CAF1 22413 117 - 1 AGUUGCCC---GGCCUCAAUCGAUACCUAUCGACAAGCGUGAGCAGCCCUGUCUGUGCAUGUGCUGCCAUUCGAUUCGAUGACCAGCACUCGGUACACACGUUUUUUGGCUCUGUACAAA ....(((.---(((.....(((((....)))))...((....)).)))..((.(((((..((((((.((((......))))..))))))...))))).)).......))).......... ( -35.10) >consensus AUAUGUAC___UGCCCCAAUCGAUACCUAUCGAUGAGCGUGGGCAGCCCUGUUUGUGCAUGUGCUGCCAUUCGAUUAGAUGACCAGCACUCCGUGCACACGUUUUUCGGCUUUCUAGAAA ............(((...((((((....))))))(((((((.(((....))).(((((..((((((....((.....))....))))))...))))))))))))...))).......... (-29.86 = -29.87 + 0.01)

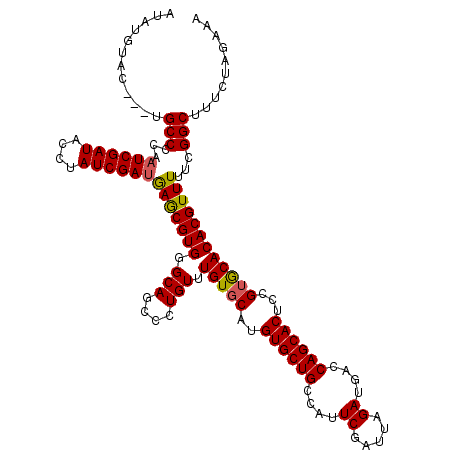

| Location | 5,266,088 – 5,266,204 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.97 |
| Mean single sequence MFE | -36.08 |
| Consensus MFE | -29.71 |
| Energy contribution | -27.50 |
| Covariance contribution | -2.21 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5266088 116 + 22224390 AUCAAAUCGAAUGGCAGCACAUGCACAAACAGGGCUGCCCACGCUCAUCGAUAGGUAUCGAUUGGGGCA---AUAGGUAUUUUCUUAGGCGGCGUUGCCACC-GAAUGUGCACAAAAGUG ......(((..(((((((.(.((......)).)((((((...((((((((((....))))))..)))).---.((((......))))))))))))))))).)-)).....(((....))) ( -35.00) >DroSim_CAF1 21405 120 + 1 UUCUAAUCGAAUGGCAGCACAUGUACAAACAGGGCUGCCCACGUUCAUCGAUAUGUAUCGAUAUGGGCAUAAGUACAUAUCUUCCAAGGCGGUGUUGCCACCUGACUGUUCCCAAAAGUG ......(((..(((((((((((((((.....(((...)))..((((((((((....)))))..)))))....)))))).(((....)))..)))))))))..)))............... ( -33.20) >DroYak_CAF1 22453 114 + 1 AUCGAAUCGAAUGGCAGCACAUGCACAGACAGGGCUGCUCACGCUUGUCGAUAGGUAUCGAUUGAGGCC---GGGCAACUCAGCUCUGGCGGCGCUGCCACC-UACUG--CGCAUAAUUG ..((.......(((((((.(.(((.(((((.(((.(((((..((((((((((....))))))..)))).---))))).))).).)))))))).)))))))..-.....--))........ ( -40.04) >consensus AUCAAAUCGAAUGGCAGCACAUGCACAAACAGGGCUGCCCACGCUCAUCGAUAGGUAUCGAUUGGGGCA___GUACAUAUCUUCUAAGGCGGCGUUGCCACC_GACUGU_CACAAAAGUG ...........(((((((.(.((......)).)((((((...((((((((((....))))))..))))....((....)).......))))))))))))).................... (-29.71 = -27.50 + -2.21)



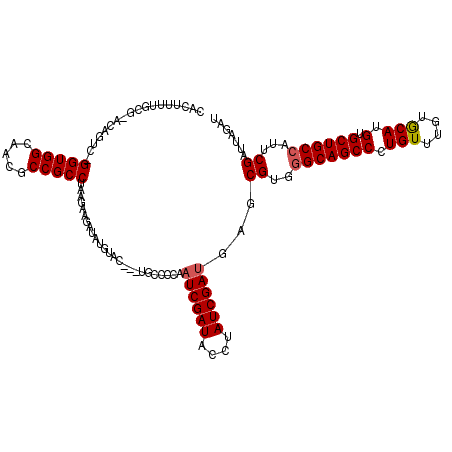
| Location | 5,266,088 – 5,266,204 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.97 |
| Mean single sequence MFE | -38.13 |
| Consensus MFE | -25.59 |
| Energy contribution | -26.03 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5266088 116 - 22224390 CACUUUUGUGCACAUUC-GGUGGCAACGCCGCCUAAGAAAAUACCUAU---UGCCCCAAUCGAUACCUAUCGAUGAGCGUGGGCAGCCCUGUUUGUGCAUGUGCUGCCAUUCGAUUUGAU (((....)))(((....-((((((...))))))...............---.((....((((((....))))))..)))))(((((((.(((....))).).))))))............ ( -35.00) >DroSim_CAF1 21405 120 - 1 CACUUUUGGGAACAGUCAGGUGGCAACACCGCCUUGGAAGAUAUGUACUUAUGCCCAUAUCGAUACAUAUCGAUGAACGUGGGCAGCCCUGUUUGUACAUGUGCUGCCAUUCGAUUAGAA ..((..(.(((.......((((....))))....(((.((((((((((...(((((((((((((....))))))....))))))).........)))))))).)).)))))).)..)).. ( -38.80) >DroYak_CAF1 22453 114 - 1 CAAUUAUGCG--CAGUA-GGUGGCAGCGCCGCCAGAGCUGAGUUGCCC---GGCCUCAAUCGAUACCUAUCGACAAGCGUGAGCAGCCCUGUCUGUGCAUGUGCUGCCAUUCGAUUCGAU ........((--...(.-.((((((((((.((((((((......))..---(((.....(((((....)))))...((....)).)))...)))).))..))))))))))..)...)).. ( -40.60) >consensus CACUUUUGCG_ACAGUC_GGUGGCAACGCCGCCUAAGAAGAUAUGUAC___UGCCCCAAUCGAUACCUAUCGAUGAGCGUGGGCAGCCCUGUUUGUGCAUGUGCUGCCAUUCGAUUAGAU ..................(((((.....))))).........................((((((....))))))...((..(((((((.(((....))).).))))))...))....... (-25.59 = -26.03 + 0.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:21 2006