
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,264,807 – 5,264,908 |
| Length | 101 |
| Max. P | 0.996524 |

| Location | 5,264,807 – 5,264,908 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 92.38 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -30.01 |
| Energy contribution | -30.23 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.82 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996524 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
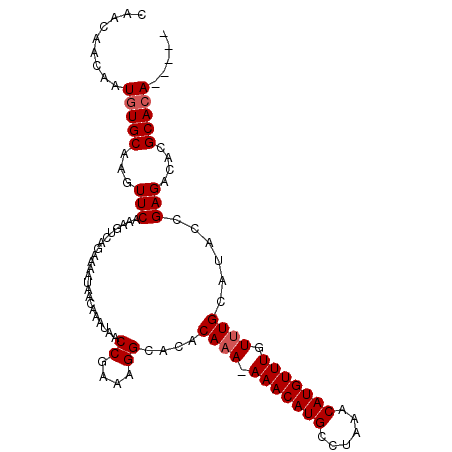
>X_DroMel_CAF1 5264807 101 + 22224390 -----UCUGCGUGUCUCGGCAUGCAAACAAACAUGUUUAGGCAUGUUU-UUUGUGUGCCUUUCGGUUAUUUGUUAUUUUCUGACUUUGAACUUGCACAUUGUUGUUG -----.....((((...((((((((((.((((((((....))))))))-))))))))))....((((....((((.....))))....)))).)))).......... ( -28.80) >DroSim_CAF1 20126 102 + 1 -----UGUGCGUGUCUCGGUAUGCAAACAAACAUGUUUAGGCAUGUUUUUUUGUGUGCCUUUCGGUUAUUUGUUAUUUUCUGACUUUGAACUUGCACAUUGUUGUUG -----((((((.((.((((((((((((.((((((((....)))))))).))))))))))..((((..(........)..))))....)))).))))))......... ( -31.20) >DroYak_CAF1 21183 106 + 1 GUGUCUGUGCGUGUCUCGGUAUGCAAACAAACAUGUUUAGGCAUGUUU-UGUGUAUGCCUUUUGGUUAUUUGUUAUUUUUUGACUUUGAACUUGCACAUUGUUGUUG .....((((((......((((((((...((((((((....))))))))-..))))))))....((((....((((.....))))....))))))))))......... ( -30.10) >consensus _____UGUGCGUGUCUCGGUAUGCAAACAAACAUGUUUAGGCAUGUUU_UUUGUGUGCCUUUCGGUUAUUUGUUAUUUUCUGACUUUGAACUUGCACAUUGUUGUUG .....((((((......((((((((((.((((((((....)))))))).))))))))))....((((....((((.....))))....))))))))))......... (-30.01 = -30.23 + 0.23)



| Location | 5,264,807 – 5,264,908 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 92.38 |
| Mean single sequence MFE | -18.19 |
| Consensus MFE | -15.93 |
| Energy contribution | -16.60 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.931950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5264807 101 - 22224390 CAACAACAAUGUGCAAGUUCAAAGUCAGAAAAUAACAAAUAACCGAAAGGCACACAAA-AAACAUGCCUAAACAUGUUUGUUUGCAUGCCGAGACACGCAGA----- ...........(((...(((.......)))..................((((..((((-(((((((......))))))).))))..)))).......)))..----- ( -17.90) >DroSim_CAF1 20126 102 - 1 CAACAACAAUGUGCAAGUUCAAAGUCAGAAAAUAACAAAUAACCGAAAGGCACACAAAAAAACAUGCCUAAACAUGUUUGUUUGCAUACCGAGACACGCACA----- .........(((((...(((......................((....))....((((.(((((((......))))))).))))......)))....)))))----- ( -19.60) >DroYak_CAF1 21183 106 - 1 CAACAACAAUGUGCAAGUUCAAAGUCAAAAAAUAACAAAUAACCAAAAGGCAUACACA-AAACAUGCCUAAACAUGUUUGUUUGCAUACCGAGACACGCACAGACAC .........(((((..(((.....((.......((((((((......((((((.....-....)))))).....))))))))........)))))..)))))..... ( -17.06) >consensus CAACAACAAUGUGCAAGUUCAAAGUCAGAAAAUAACAAAUAACCGAAAGGCACACAAA_AAACAUGCCUAAACAUGUUUGUUUGCAUACCGAGACACGCACA_____ .........(((((...(((......................((....))....((((.(((((((......))))))).))))......)))....)))))..... (-15.93 = -16.60 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:18 2006