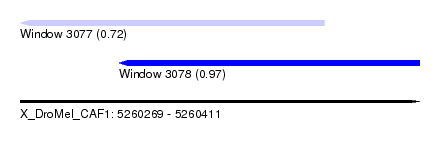
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,260,269 – 5,260,411 |
| Length | 142 |
| Max. P | 0.973539 |

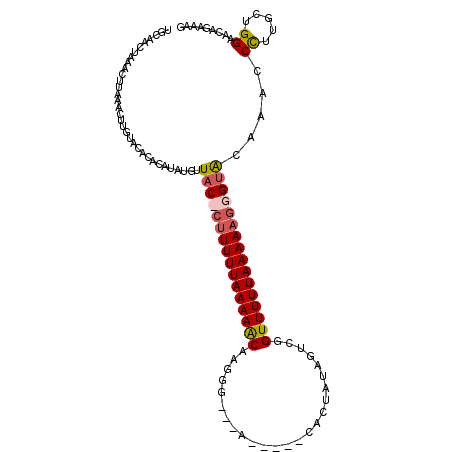
| Location | 5,260,269 – 5,260,377 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 84.36 |
| Mean single sequence MFE | -28.07 |
| Consensus MFE | -15.17 |
| Energy contribution | -16.73 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5260269 108 - 22224390 AC-CUUUUUAAAAACAAGCGGAUG-----CGCUAUCGUCUGUUUUUAAAAAGGGGACAAACCCUUGUUGGAACAGAAAGUUUCUAACUCUUGUUUCUAAUUGCAGAAUGUAAUC .(-(((((((((((((..(((...-----.....)))..)))))))))))))).(((((....)))))(((((((..(((.....))).)))))))..(((((.....))))). ( -27.90) >DroSec_CAF1 15936 106 - 1 ACCCUUUUUAAAAACAAGGG---A-----CACUAUAGUCGGUUUUUAAAAAGGGUACCAAUCCUUGCUGGAACAGAAAGUUUCUAACUCUUGUUUCUAAUUGCAGAAUGUAAUC (((((((((((((((....(---(-----(......))).)))))))))))))))........((((.(((((((..(((.....))).))))))).....))))......... ( -28.00) >DroSim_CAF1 15600 106 - 1 ACCCUUUUUAAAAACAAGGG---A-----CAUUAUAGUCGGUUUUUAAAAAGGGUACCAAUCCUUGCUGGAACGGAAAGUUUCUAACUCUUGUUUCUAAUUGCAGAAUGUAAUC (((((((((((((((....(---(-----(......))).)))))))))))))))........((((.(((((((..(((.....))).))))))).....))))......... ( -27.20) >DroEre_CAF1 16645 111 - 1 AC-CUUUUUAAAAGCAAAGGGGAGCACCCCACUCCCGUCUGUUUUUAAAA--GGUGCAAACCUUUUCUGGUACAGAAAAUUUCUAACUCUUGUUUCUAAUUGCAGAAUGUAAUC ..-(((((((((((((.(.(((((.......))))).).)))))))))))--))(((((...(((((((...))))))).....(((....))).....))))).......... ( -29.20) >consensus AC_CUUUUUAAAAACAAGGG___A_____CACUAUAGUCGGUUUUUAAAAAGGGUACAAACCCUUGCUGGAACAGAAAGUUUCUAACUCUUGUUUCUAAUUGCAGAAUGUAAUC .......(((.(((((((((.................((((((((....((((((....))))))...))))))))..........))))))))).)))((((.....)))).. (-15.17 = -16.73 + 1.56)

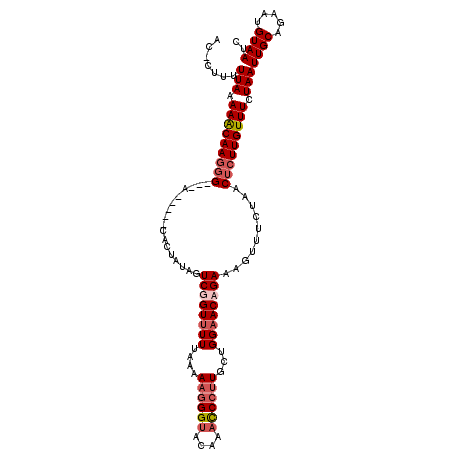
| Location | 5,260,304 – 5,260,411 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.24 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -11.80 |
| Energy contribution | -12.48 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.64 |
| Structure conservation index | 0.42 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5260304 107 - 22224390 UGCAACUAAACUUAAACUUGUACACACAUAUGUUAC-CUUUUUAAAAACAAGCGGAUG-----CGCUAUCGUCUGUUUUUAAAAAGGGGACAAACCCUUGUUGGAACAGAAAG ..((((............(((....)))..(((..(-(((((((((((((..(((...-----.....)))..))))))))))))))..))).......)))).......... ( -25.90) >DroSec_CAF1 15971 99 - 1 UGCAACUAAACAU------AUACACACAUAUGUUACCCUUUUUAAAAACAAGGG---A-----CACUAUAGUCGGUUUUUAAAAAGGGUACCAAUCCUUGCUGGAACAGAAAG .((((...(((((------((......)))))))(((((((((((((((....(---(-----(......))).)))))))))))))))........))))............ ( -27.70) >DroSim_CAF1 15635 105 - 1 UGCAACUAAACUUAAACUUGUACACACAUAUGUUACCCUUUUUAAAAACAAGGG---A-----CAUUAUAGUCGGUUUUUAAAAAGGGUACCAAUCCUUGCUGGAACGGAAAG .................................((((((((((((((((....(---(-----(......))).))))))))))))))))((..(((.....)))..)).... ( -26.20) >DroEre_CAF1 16680 108 - 1 UGCAACUUUACUUCGACUUGUAA--ACAUAAGUUAC-CUUUUUAAAAGCAAAGGGGAGCACCCCACUCCCGUCUGUUUUUAAAA--GGUGCAAACCUUUUCUGGUACAGAAAA ((((..........(((((((..--..)))))))..-(((((((((((((.(.(((((.......))))).).)))))))))))--)))))).....((((((...)))))). ( -32.20) >consensus UGCAACUAAACUUAAACUUGUACACACAUAUGUUAC_CUUUUUAAAAACAAGGG___A_____CACUAUAGUCGGUUUUUAAAAAGGGUACAAACCCUUGCUGGAACAGAAAG .................................((((((((((((((((.........................)))))))))))))))).....((.....))......... (-11.80 = -12.48 + 0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:14 2006