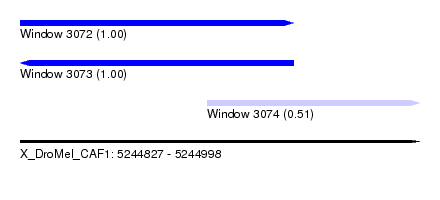
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,244,827 – 5,244,998 |
| Length | 171 |
| Max. P | 0.999964 |

| Location | 5,244,827 – 5,244,944 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.31 |
| Mean single sequence MFE | -23.10 |
| Consensus MFE | -22.97 |
| Energy contribution | -23.14 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.996413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5244827 117 + 22224390 UGCCACCUAUAUCUAGUAGAACACCACAUUUAGUGCCUGAUACUCACAUGCCAUCAAAUGGAUCUGCAGUUAAUAGCUACGCAUCUUCCAACAAAUACGGCAGCUGAAUAUCAGA---AA ......((((.....)))).....(((.....))).((((((.(((..((((......((((..((((((.....)))..)))...))))........))))..))).)))))).---.. ( -23.84) >DroVir_CAF1 1288 117 + 1 UGCCACCUAUAUCUAGUAGAACACCACAUUUAGUGCCUGAUACUCACAUGCCAUCAAAUGGAUCUGCAGUUAAUAGCUACGCAUCUUCCAACAAAUACGGCAGCUGAAUAUCCGA---AA ......((((.....))))........((((((((((((((.(......)..))))..((((..((((((.....)))..)))...))))........)))).))))))......---.. ( -19.40) >DroPse_CAF1 1046 120 + 1 UGCCACCUAUAUCUAGUAGAACACCACAUUUAGUGCCUGAUACUCACAUGCCAUCAAAUGGAUCUGCAGUUAAUAGCUACGCAUCUUCCAACAAAUACGGCAGUUGAAUAUCAGAGAGAA ......((((.....)))).....(((.....))).((((((.(((..((((......((((..((((((.....)))..)))...))))........))))..))).))))))...... ( -23.84) >DroYak_CAF1 1007 117 + 1 UGCCACCUAUAUCUAGUAGAACACCACAUUUAGUGCCUGAUACUCACAUGCCAUCAAAUGGAUCUGCAGUUAAUAGCUACGCAUCUUCCAACAAAUACGGCAGCUGAAUAUCAGA---AA ......((((.....)))).....(((.....))).((((((.(((..((((......((((..((((((.....)))..)))...))))........))))..))).)))))).---.. ( -23.84) >DroMoj_CAF1 1554 117 + 1 UGCCACCUAUAUCUAGUAGAACACCACAUUUAGUGCCUGAUACUCACAUGCCAUCAAAUGGAUCUGCAGUUAAUAGCUACGCAUCUUCCAACAAAUACGGCAGCUGAAUAUCAGA---CA ......((((.....)))).....(((.....))).((((((.(((..((((......((((..((((((.....)))..)))...))))........))))..))).)))))).---.. ( -23.84) >DroAna_CAF1 1032 117 + 1 UGCCACCUAUAUCUAGUAGAACACCACAUUUAGUGCCUGAUACUCACAUGCCAUCAAAUGGAUCUGCAGUUAAUAGCUACGCAUCUUCCAACAAAUACGGCAGCUGAAUAUCAGA---AA ......((((.....)))).....(((.....))).((((((.(((..((((......((((..((((((.....)))..)))...))))........))))..))).)))))).---.. ( -23.84) >consensus UGCCACCUAUAUCUAGUAGAACACCACAUUUAGUGCCUGAUACUCACAUGCCAUCAAAUGGAUCUGCAGUUAAUAGCUACGCAUCUUCCAACAAAUACGGCAGCUGAAUAUCAGA___AA ......((((.....)))).....(((.....))).((((((.(((..((((......((((..((((((.....)))..)))...))))........))))..))).))))))...... (-22.97 = -23.14 + 0.17)

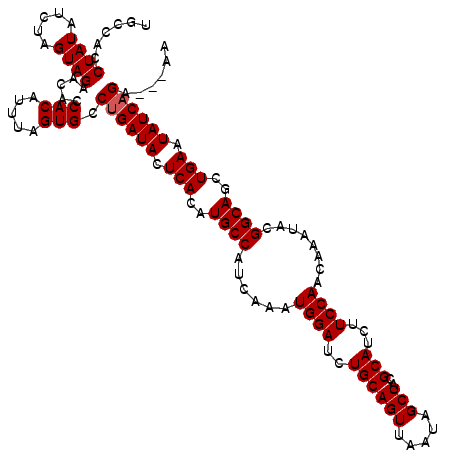
| Location | 5,244,827 – 5,244,944 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.31 |
| Mean single sequence MFE | -37.13 |
| Consensus MFE | -36.47 |
| Energy contribution | -36.63 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.98 |
| SVM decision value | 4.94 |
| SVM RNA-class probability | 0.999964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5244827 117 - 22224390 UU---UCUGAUAUUCAGCUGCCGUAUUUGUUGGAAGAUGCGUAGCUAUUAACUGCAGAUCCAUUUGAUGGCAUGUGAGUAUCAGGCACUAAAUGUGGUGUUCUACUAGAUAUAGGUGGCA ..---.((((((((((..((((((......((((...(((((........)).)))..))))....))))))..))))))))))((.(((.((.(((((...))))).)).)))...)). ( -37.30) >DroVir_CAF1 1288 117 - 1 UU---UCGGAUAUUCAGCUGCCGUAUUUGUUGGAAGAUGCGUAGCUAUUAACUGCAGAUCCAUUUGAUGGCAUGUGAGUAUCAGGCACUAAAUGUGGUGUUCUACUAGAUAUAGGUGGCA ..---...........((..(((((((((.((((.(((((...((((((((.((......)).))))))))......)))))..((((((....)))))))))).))))))).))..)). ( -34.20) >DroPse_CAF1 1046 120 - 1 UUCUCUCUGAUAUUCAACUGCCGUAUUUGUUGGAAGAUGCGUAGCUAUUAACUGCAGAUCCAUUUGAUGGCAUGUGAGUAUCAGGCACUAAAUGUGGUGUUCUACUAGAUAUAGGUGGCA ......((((((((((..((((((......((((...(((((........)).)))..))))....))))))..))))))))))((.(((.((.(((((...))))).)).)))...)). ( -37.50) >DroYak_CAF1 1007 117 - 1 UU---UCUGAUAUUCAGCUGCCGUAUUUGUUGGAAGAUGCGUAGCUAUUAACUGCAGAUCCAUUUGAUGGCAUGUGAGUAUCAGGCACUAAAUGUGGUGUUCUACUAGAUAUAGGUGGCA ..---.((((((((((..((((((......((((...(((((........)).)))..))))....))))))..))))))))))((.(((.((.(((((...))))).)).)))...)). ( -37.30) >DroMoj_CAF1 1554 117 - 1 UG---UCUGAUAUUCAGCUGCCGUAUUUGUUGGAAGAUGCGUAGCUAUUAACUGCAGAUCCAUUUGAUGGCAUGUGAGUAUCAGGCACUAAAUGUGGUGUUCUACUAGAUAUAGGUGGCA ((---(((((((((((..((((((......((((...(((((........)).)))..))))....))))))..)))))))))))))(((.((.(((((...))))).)).)))...... ( -39.20) >DroAna_CAF1 1032 117 - 1 UU---UCUGAUAUUCAGCUGCCGUAUUUGUUGGAAGAUGCGUAGCUAUUAACUGCAGAUCCAUUUGAUGGCAUGUGAGUAUCAGGCACUAAAUGUGGUGUUCUACUAGAUAUAGGUGGCA ..---.((((((((((..((((((......((((...(((((........)).)))..))))....))))))..))))))))))((.(((.((.(((((...))))).)).)))...)). ( -37.30) >consensus UU___UCUGAUAUUCAGCUGCCGUAUUUGUUGGAAGAUGCGUAGCUAUUAACUGCAGAUCCAUUUGAUGGCAUGUGAGUAUCAGGCACUAAAUGUGGUGUUCUACUAGAUAUAGGUGGCA ......((((((((((..((((((......((((...(((((........)).)))..))))....))))))..))))))))))((.(((.((.(((((...))))).)).)))...)). (-36.47 = -36.63 + 0.17)



| Location | 5,244,907 – 5,244,998 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.83 |
| Mean single sequence MFE | -20.19 |
| Consensus MFE | -16.36 |
| Energy contribution | -16.16 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5244907 91 + 22224390 GCAUCUUCCAACAAAUACGGCAGCUGAAUAUCAGA---AAGAGAGCGCUCCAACUCCGUGAUUAACAUCGAGAUGCUAACAAUAAGGCCGGAUC-------------------------- (((((((........(((((.(((((.....))).---..(((....)))...))))))).........)))))))..................-------------------------- ( -17.73) >DroVir_CAF1 1368 91 + 1 GCAUCUUCCAACAAAUACGGCAGCUGAAUAUCCGA---AAGAGAGCGCUCCAACUCCGUGAUUAACAUCGAGAUGCAUAUAUUAAAACCAGCUA-------------------------- (((((((........(((((.((.((....(((..---..).))......)).))))))).........)))))))..................-------------------------- ( -18.13) >DroPse_CAF1 1126 89 + 1 GCAUCUUCCAACAAAUACGGCAGUUGAAUAUCAGAGAGAAGAGAGCGCUCCAGUUCCGUGAUUAACAUCGAGAUGCUACCAUUAAAGCC------------------------------- (((((((.((((..........))))...((((..(((..(((....)))...)))..)))).......))))))).............------------------------------- ( -16.00) >DroGri_CAF1 1114 91 + 1 GCAUCUUCCAACAAAUACGGCAGCUGAAUAUCAGA---AAGAGAGCGCUCCAACUCCGUGAUUAACAUCGAGAUGCAUACAAUAAGGCCAACUA-------------------------- (((((((........(((((.(((((.....))).---..(((....)))...))))))).........)))))))..................-------------------------- ( -18.23) >DroMoj_CAF1 1634 88 + 1 GCAUCUUCCAACAAAUACGGCAGCUGAAUAUCAGA---CAGAGAGCGCUCCAACUCCGUGAUUAACAUUGAGAUGCAUCCAUUACGACCCA----------------------------- (((((((........(((((.(((((.....))).---..(((....)))...))))))).........)))))))...............----------------------------- ( -18.23) >DroAna_CAF1 1112 117 + 1 GCAUCUUCCAACAAAUACGGCAGCUGAAUAUCAGA---AAGAGAGCGCUCCACUUCCGUGAUUAACAUCGAGAUGCUAACAAUAAGGUCGGAUCUGGAUCCCACAAUCCGGGAUUCAGGA (((((((........(((((.(((((.....))).---..(((....)))..)).))))).........)))))))................((((((((((.......)))))))))). ( -32.83) >consensus GCAUCUUCCAACAAAUACGGCAGCUGAAUAUCAGA___AAGAGAGCGCUCCAACUCCGUGAUUAACAUCGAGAUGCAAACAAUAAGGCCAG_U___________________________ (((((((........(((((.(((((.....)))......(((....)))...))))))).........)))))))............................................ (-16.36 = -16.16 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:11 2006