
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,239,230 – 5,239,440 |
| Length | 210 |
| Max. P | 0.999917 |

| Location | 5,239,230 – 5,239,327 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 81.62 |
| Mean single sequence MFE | -16.91 |
| Consensus MFE | -12.44 |
| Energy contribution | -12.63 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
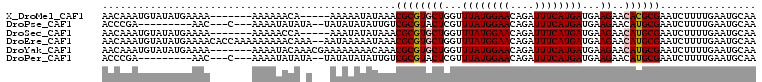
>X_DroMel_CAF1 5239230 97 + 22224390 AACAAAUGUAUAUGAAAA-------AAAAAACA-----AAAAAUAUAAACGCGUGCUGGUUUAUGGAACAGAUUUCAUGAUGAAGAACACGCGAAUCUUUUGAAUGCAA ......(((((.......-------........-----...........((((((((...((((((((....))))))))...))..))))))..((....))))))). ( -18.10) >DroPse_CAF1 16217 92 + 1 ACCCGA---------AAC---C---AAAAUAUAUA--UAUAUAUAUUGUCGCGUACUCGUUUAUGGAACAGAUUUCAUGAUGAAGAACAUGCGAAUCUUUUGAAUGCAA ...(((---------((.---.---..(((((((.--...))))))).((((((..((..((((((((....))))))))....))..))))))...)))))....... ( -16.70) >DroSec_CAF1 12411 97 + 1 AACAAAUGUAUAUGAAAA-------AAAAACCA-----AAAUAUAUAAACGCGUGCUGGUUUAUGGAACAGAUUUCAUGAUGAAGAACAUGCGAAUCUUUUGAAUGCAA ..(((((((((((.....-------........-----..)))))))..((((((((...((((((((....))))))))...))..)))))).....))))....... ( -17.16) >DroEre_CAF1 12457 107 + 1 AACAAAUGUAUAUGAAAACACCAAAAAAAAACAAA--AAUAAAAAUAAACGCGUGCUGGUUUAUGGAACAGAUUUCAUGAUGAAGAACAUGCGAAUCUUUUGAAUGCAA ......(((((........................--............((((((((...((((((((....))))))))...))..))))))..((....))))))). ( -16.30) >DroYak_CAF1 12605 102 + 1 AACAAAUGUAUAUGAAAA-------AAAAUACAAACGAAAAAAAACAAACGCGUGCUGGUUUAUGGAACAGAUUUCAUGAUGAAGAACAUGCGAAUCUUUUGAAUGCAA ..(((((((((.(.....-------.).)))))................((((((((...((((((((....))))))))...))..)))))).....))))....... ( -16.50) >DroPer_CAF1 8079 92 + 1 ACCCGA---------AAC---C---AAAAUAUAUA--UAUAUAUAUUGUCGCGUACUCGUUUAUGGAACAGAUUUCAUGAUGAAGAACAUGCGAAUCUUUUGAAUGCAA ...(((---------((.---.---..(((((((.--...))))))).((((((..((..((((((((....))))))))....))..))))))...)))))....... ( -16.70) >consensus AACAAAUGUAUAUGAAAA_______AAAAAACA_A___AAAAAUAUAAACGCGUGCUGGUUUAUGGAACAGAUUUCAUGAUGAAGAACAUGCGAAUCUUUUGAAUGCAA .................................................((((((((...((((((((....))))))))...))..))))))................ (-12.44 = -12.63 + 0.19)

| Location | 5,239,287 – 5,239,407 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -18.92 |
| Consensus MFE | -18.69 |
| Energy contribution | -18.50 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5239287 120 - 22224390 AGCGAUACCACGUCCACAGUACGUAUUCGGAACAAUAUCACCGAAACCAACACUCAAAAAUGUUAUCGCUAUCAGCCACAUUGCAUUCAAAAGAUUCGCGUGUUCUUCAUCAUGAAAUCU (((((((.(((((.......)))).(((((..........)))))................).)))))))....((......)).......(((((..((((........)))).))))) ( -19.90) >DroVir_CAF1 18009 120 - 1 AGCGAUACCACGUCCACAGUACGUAUUCGGAACAAUAUCACCGAAACCAACACUCAAAAAUGUUAUCGCUAUCAGCCACAUUGCAUUUAAAAGAUUCGCAUGCUCUUCAUCAUGAAAUCU (((((((.(((((.......)))).(((((..........)))))................).)))))))....((......)).......(((((..((((........)))).))))) ( -19.30) >DroPse_CAF1 16269 120 - 1 AGCGAUACCACGUCCACAGUACGUAUUCGGAACAAUAUCACCGAAACCAACACUCAAAAAUGUUAUUGCUAUCAGCCACAUUGCAUUCAAAAGAUUCGCAUGUUCUUCAUCAUGAAAUCU (((((((.(((((.......)))).(((((..........)))))................).)))))))....((......)).......(((((..((((........)))).))))) ( -18.20) >DroGri_CAF1 16894 120 - 1 AGCGAUACCACGUCCACAGUACGUAUUCGGAACAAUAUCACCGAAACCAACACUCAAAAAUGUUAUCGCUAUCAGCCACAUUGCAUUUAAAAGAUUCGCAUGCUCUUCGUCAUGAAAUCU (((((((.(((((.......)))).(((((..........)))))................).)))))))....((......)).......(((((..(((((.....).)))).))))) ( -19.80) >DroMoj_CAF1 20014 120 - 1 AGCGAUACCACGUCCACAGUACGUAUUCGGAACAAUAUCCCCGAAACCAACACUCAAAAAUGUUAUCGCUAUCAGCCACAUUGCAUUUAAUAAAUUCGCAUGUUCUUCAUCAUGAAAUCU (((((((.(((((.......)))).(((((..........)))))................).)))))))....((......))..............((((........))))...... ( -18.10) >DroPer_CAF1 8131 120 - 1 AGCGAUACCACGUCCACAGUACGUAUUCGGAACAAUAUCACCGAAACCAACACUCAAAAAUGUUAUUGCUAUCAGCCACAUUGCAUUCAAAAGAUUCGCAUGUUCUUCAUCAUGAAAUCU (((((((.(((((.......)))).(((((..........)))))................).)))))))....((......)).......(((((..((((........)))).))))) ( -18.20) >consensus AGCGAUACCACGUCCACAGUACGUAUUCGGAACAAUAUCACCGAAACCAACACUCAAAAAUGUUAUCGCUAUCAGCCACAUUGCAUUCAAAAGAUUCGCAUGUUCUUCAUCAUGAAAUCU (((((((.(((((.......)))).(((((..........)))))................).)))))))....((......)).......(((((..((((........)))).))))) (-18.69 = -18.50 + -0.19)


| Location | 5,239,327 – 5,239,440 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.70 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -29.52 |
| Energy contribution | -29.85 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.987863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5239327 113 + 22224390 UGUGGCUGAUAGCGAUAACAUUUUUGAGUGUUGGUUUCGGUGAUAUUGUUCCGAAUACGUACUGUGGACGUGGUAUCGCUGUCAGUACAGGAAUAAUGGUGAGUUUUUAACAA------- ((((.((((((((((((.(((.(..(((((.....(((((..........)))))....)))).)..).))).))))))))))))))))........................------- ( -31.50) >DroVir_CAF1 18049 114 + 1 UGUGGCUGAUAGCGAUAACAUUUUUGAGUGUUGGUUUCGGUGAUAUUGUUCCGAAUACGUACUGUGGACGUGGUAUCGCUGUCAGUACAGGAAUAAUGGUGAGUUUUUAACAAA------ ((((.((((((((((((.(((.(..(((((.....(((((..........)))))....)))).)..).))).)))))))))))))))).........................------ ( -31.50) >DroPse_CAF1 16309 120 + 1 UGUGGCUGAUAGCAAUAACAUUUUUGAGUGUUGGUUUCGGUGAUAUUGUUCCGAAUACGUACUGUGGACGUGGUAUCGCUGUCAGUACAGGAAUAAUGGUGAGUUUUUAACAAACAACUA ((((.((((((((..(((((((....)))))))(((((((..........))))).))((((..(....)..)))).)))))))))))).......((((..((((.....)))).)))) ( -29.80) >DroWil_CAF1 18382 115 + 1 UGUGGCUGAUAGCGAUAACAUUUUUGAGUGUUGGUUUCGGUGAUAUUGUUCCGAAUACGUACUGUGGACGUGGUAUCGCUGUCAGUACAGGAAUAAUGGUGAGUUUUUA-CAA----UAA ((((.((((((((((((.(((.(..(((((.....(((((..........)))))....)))).)..).))).)))))))))))))))).........((((....)))-)..----... ( -31.90) >DroMoj_CAF1 20054 114 + 1 UGUGGCUGAUAGCGAUAACAUUUUUGAGUGUUGGUUUCGGGGAUAUUGUUCCGAAUACGUACUGUGGACGUGGUAUCGCUGUCAGUACAGGAAUAAUGGUGAGUUUUUAACCUU------ ((((.((((((((((((.(((.(..(((((.....(((((((......)))))))....)))).)..).))).))))))))))))))))........(((.........)))..------ ( -35.10) >DroPer_CAF1 8171 120 + 1 UGUGGCUGAUAGCAAUAACAUUUUUGAGUGUUGGUUUCGGUGAUAUUGUUCCGAAUACGUACUGUGGACGUGGUAUCGCUGUCAGUACAGGAAUAAUGGUGAGUUUUUAACAAACAACUA ((((.((((((((..(((((((....)))))))(((((((..........))))).))((((..(....)..)))).)))))))))))).......((((..((((.....)))).)))) ( -29.80) >consensus UGUGGCUGAUAGCGAUAACAUUUUUGAGUGUUGGUUUCGGUGAUAUUGUUCCGAAUACGUACUGUGGACGUGGUAUCGCUGUCAGUACAGGAAUAAUGGUGAGUUUUUAACAAA______ ((((.((((((((((((.(((.(..(((((.....(((((..........)))))....)))).)..).))).))))))))))))))))............................... (-29.52 = -29.85 + 0.33)



| Location | 5,239,327 – 5,239,440 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.70 |
| Mean single sequence MFE | -20.48 |
| Consensus MFE | -20.19 |
| Energy contribution | -19.97 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.54 |
| SVM RNA-class probability | 0.999917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5239327 113 - 22224390 -------UUGUUAAAAACUCACCAUUAUUCCUGUACUGACAGCGAUACCACGUCCACAGUACGUAUUCGGAACAAUAUCACCGAAACCAACACUCAAAAAUGUUAUCGCUAUCAGCCACA -------.........................((.((((.(((((((.(((((.......)))).(((((..........)))))................).))))))).))))..)). ( -20.60) >DroVir_CAF1 18049 114 - 1 ------UUUGUUAAAAACUCACCAUUAUUCCUGUACUGACAGCGAUACCACGUCCACAGUACGUAUUCGGAACAAUAUCACCGAAACCAACACUCAAAAAUGUUAUCGCUAUCAGCCACA ------..........................((.((((.(((((((.(((((.......)))).(((((..........)))))................).))))))).))))..)). ( -20.60) >DroPse_CAF1 16309 120 - 1 UAGUUGUUUGUUAAAAACUCACCAUUAUUCCUGUACUGACAGCGAUACCACGUCCACAGUACGUAUUCGGAACAAUAUCACCGAAACCAACACUCAAAAAUGUUAUUGCUAUCAGCCACA .....((((.....))))..............((.((((.(((((((.(((((.......)))).(((((..........)))))................).))))))).))))..)). ( -19.50) >DroWil_CAF1 18382 115 - 1 UUA----UUG-UAAAAACUCACCAUUAUUCCUGUACUGACAGCGAUACCACGUCCACAGUACGUAUUCGGAACAAUAUCACCGAAACCAACACUCAAAAAUGUUAUCGCUAUCAGCCACA ...----...-.....................((.((((.(((((((.(((((.......)))).(((((..........)))))................).))))))).))))..)). ( -20.60) >DroMoj_CAF1 20054 114 - 1 ------AAGGUUAAAAACUCACCAUUAUUCCUGUACUGACAGCGAUACCACGUCCACAGUACGUAUUCGGAACAAUAUCCCCGAAACCAACACUCAAAAAUGUUAUCGCUAUCAGCCACA ------..(((.........))).........((.((((.(((((((.(((((.......)))).(((((..........)))))................).))))))).))))..)). ( -22.10) >DroPer_CAF1 8171 120 - 1 UAGUUGUUUGUUAAAAACUCACCAUUAUUCCUGUACUGACAGCGAUACCACGUCCACAGUACGUAUUCGGAACAAUAUCACCGAAACCAACACUCAAAAAUGUUAUUGCUAUCAGCCACA .....((((.....))))..............((.((((.(((((((.(((((.......)))).(((((..........)))))................).))))))).))))..)). ( -19.50) >consensus ______UUUGUUAAAAACUCACCAUUAUUCCUGUACUGACAGCGAUACCACGUCCACAGUACGUAUUCGGAACAAUAUCACCGAAACCAACACUCAAAAAUGUUAUCGCUAUCAGCCACA ................................((.((((.(((((((.(((((.......)))).(((((..........)))))................).))))))).))))..)). (-20.19 = -19.97 + -0.22)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:06 2006