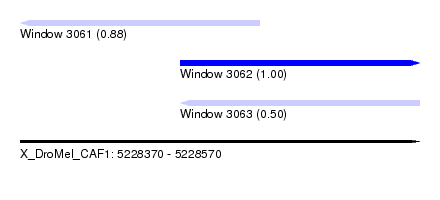
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,228,370 – 5,228,570 |
| Length | 200 |
| Max. P | 0.995661 |

| Location | 5,228,370 – 5,228,490 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.61 |
| Mean single sequence MFE | -29.04 |
| Consensus MFE | -28.02 |
| Energy contribution | -28.13 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
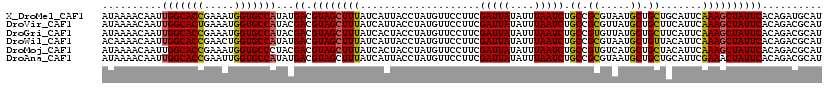
>X_DroMel_CAF1 5228370 120 - 22224390 GCUACGUCAUAUGGCACCAUUUCGGUGCCAAUUGUUUUAUUCUUAUUGGCCAAUUUCGUCGUCCACUGGAAAUAGUAUUCGCCAGGAAUUGGAUGUAUAGCGCAUAUAAAAAGUUCUAAC ((((...(((.(((((((.....)))))))...................((((((((..((...((((....))))...))...)))))))))))..))))................... ( -30.50) >DroVir_CAF1 7957 120 - 1 GCUACGUCGUAUGGCACCAUUUCAGUGCCAAUUGUUUUAUUUUUAUUGGCCAAUUUCGUCGUCCACUGGAAAUAGUAUUCACCAGGAAUUGGAUGUAUAGCGCAUAUAAAAAGUUCUAAA ((((...(((.((((((.......))))))...................((((((((.......((((....))))........)))))))))))..))))................... ( -23.76) >DroGri_CAF1 7579 120 - 1 GCUACGUCGUAUGGCACCAUUUCGGUGCCAAUUGUUUUAUUUUUAUUGGCCAAUUUUGUCGUCCACUGGAAAUAGUAUUCGCCAGGAAUUGGAUGUAUAGCGCAUAUAAAAAGUUCUAAC ((((((((...(((((((.....)))))))...............(((((.(((.((((..(((...))).)))).))).)))))......)))))..)))................... ( -29.90) >DroWil_CAF1 6553 120 - 1 GCUACGUCAUAUGGCACCAGUUCGGUGCCAAUUGUUUUGUUCUUAUUGGCCAAUUUCGUCGUCCACUGGAAAUAGUAUUCGCCAGGAAUUGGAUGUAUAGCGCAUAUAAAAAGUUCUAAU ((((...(((.(((((((.....)))))))...................((((((((..((...((((....))))...))...)))))))))))..))))................... ( -30.50) >DroMoj_CAF1 9550 120 - 1 GCUACGUCGUAGGGCACCAUUUCGGUGCCAAUUGUUUUAUUUUUAUUGGCCAAUUUCGUCGUCCACUGGAAAUAGUAUUCGCCAGGAAUUGGAUGUAUAGCGCAUAUAAAAAGUUCUAAC ((((...(((..((((((.....))))))....................((((((((..((...((((....))))...))...)))))))))))..))))................... ( -29.10) >DroAna_CAF1 6495 120 - 1 GCUACGUCAUAUGGCACCAAUUCGGUGCCAAUUGUUUUAUUCUUAUUGGCCAAUUUCGUCGUCCACUGGAAAUAGUAUUCGCCAGGAAUUGGAUGUAUAGCGCAUAUAAAAAGUUCUAAC ((((...(((.(((((((.....)))))))...................((((((((..((...((((....))))...))...)))))))))))..))))................... ( -30.50) >consensus GCUACGUCAUAUGGCACCAUUUCGGUGCCAAUUGUUUUAUUCUUAUUGGCCAAUUUCGUCGUCCACUGGAAAUAGUAUUCGCCAGGAAUUGGAUGUAUAGCGCAUAUAAAAAGUUCUAAC ((((...(((.(((((((.....)))))))...................((((((((..((...((((....))))...))...)))))))))))..))))................... (-28.02 = -28.13 + 0.11)
| Location | 5,228,450 – 5,228,570 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.89 |
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -24.94 |
| Energy contribution | -24.63 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
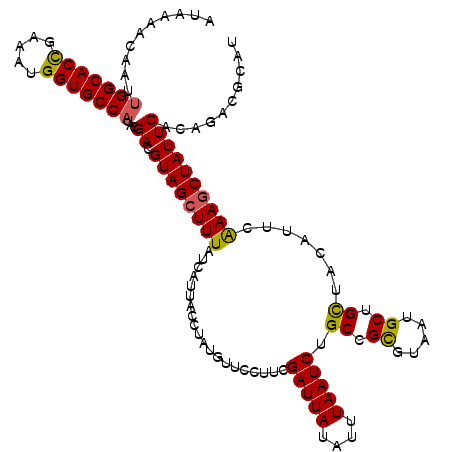
>X_DroMel_CAF1 5228450 120 + 22224390 AUAAAACAAUUGGCACCGAAAUGGUGCCAUAUGACGUAGCUUUAUCAUUACCUAUGUUCCUUCGAUUAUAUUUAAUCUGCCGCGUAAUGCUGCUGCAUUCAAAGCUAUUCACAGAUGCAU ..........(((((((.....)))))))..(((.((((((((....................(((((....)))))(((.(((......))).)))...)))))))))))......... ( -29.50) >DroVir_CAF1 8037 120 + 1 AUAAAACAAUUGGCACUGAAAUGGUGCCAUACGACGUAGCUUUAUCAUUACCUAUGUUCCUUCGAUUAUAUUUAAUCUGCCGCGUUAUGCUGCUUCAUUCAAAGCUAUUCACAGACGCAU ..........(((((((.....)))))))...(((((((.(........).))))))).....(((((....)))))....(((((.((..((((......))))....))..))))).. ( -25.70) >DroGri_CAF1 7659 120 + 1 AUAAAACAAUUGGCACCGAAAUGGUGCCAUACGACGUAGCUUUAUCACUACCUAUGUUCCUUCGAUUAUAUUUAAUCUGCCGUGUUAUGCUGCUUCAUUCAAAGCUAUUCACAGACGCAU ..........(((((((.....)))))))...((.((((((((....................(((((....))))).((.((.....)).)).......)))))))))).......... ( -25.90) >DroWil_CAF1 6633 120 + 1 ACAAAACAAUUGGCACCGAACUGGUGCCAUAUGACGUAGCUUUAUCAUUACCUAUGUUCCUUCGAUUAUAUUUAAUCUGCCGCGUAAUGCUGUUACAUUCAAAGCUAUUCACAGACGCAU ..........(((((((.....)))))))..(((.((((((((....................(((((....)))))......(((((...)))))....)))))))))))......... ( -26.10) >DroMoj_CAF1 9630 120 + 1 AUAAAACAAUUGGCACCGAAAUGGUGCCCUACGACGUAGCUUUAUCACUACCUAUGUUCCUUCGAUUAUAUUUAAUCUGCCGUGUCAUGCUGCUACAUUCAAAGCUAUUCACAGACGCAU ...........((((((.....))))))....(((((((.(........).))))))).....(((((....)))))....(((((.((..(((........)))....))..))))).. ( -24.80) >DroAna_CAF1 6575 120 + 1 AUAAAACAAUUGGCACCGAAUUGGUGCCAUAUGACGUAGCUUUAUCAUUACCUAUGUUCCUUCGAUUAUAUUUAAUCUGCCGCGUAAUGCUGCUGCAUUCGAAACUAUUCACAGACGCAU ..........(((((((.....)))))))...(((((((.(........).))))))).....(((((....)))))....(((((((((....))))).(((....)))....)))).. ( -28.20) >consensus AUAAAACAAUUGGCACCGAAAUGGUGCCAUACGACGUAGCUUUAUCAUUACCUAUGUUCCUUCGAUUAUAUUUAAUCUGCCGCGUAAUGCUGCUACAUUCAAAGCUAUUCACAGACGCAU ..........(((((((.....)))))))...((.((((((((....................(((((....))))).((.((.....)).)).......)))))))))).......... (-24.94 = -24.63 + -0.30)
| Location | 5,228,450 – 5,228,570 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.89 |
| Mean single sequence MFE | -32.08 |
| Consensus MFE | -26.19 |
| Energy contribution | -26.08 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
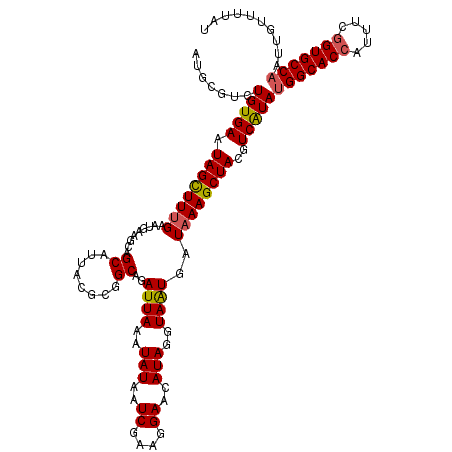
>X_DroMel_CAF1 5228450 120 - 22224390 AUGCAUCUGUGAAUAGCUUUGAAUGCAGCAGCAUUACGCGGCAGAUUAAAUAUAAUCGAAGGAACAUAGGUAAUGAUAAAGCUACGUCAUAUGGCACCAUUUCGGUGCCAAUUGUUUUAU ..(((..(((((.((((((((..(((.((........)).))).((((..(((..((....))..)))..))))..))))))))..)))))(((((((.....)))))))..)))..... ( -33.80) >DroVir_CAF1 8037 120 - 1 AUGCGUCUGUGAAUAGCUUUGAAUGAAGCAGCAUAACGCGGCAGAUUAAAUAUAAUCGAAGGAACAUAGGUAAUGAUAAAGCUACGUCGUAUGGCACCAUUUCAGUGCCAAUUGUUUUAU .(((((.((((....(((((....)))))..)))))))))...(((((....)))))...((((((......(((((........))))).((((((.......))))))..)))))).. ( -28.60) >DroGri_CAF1 7659 120 - 1 AUGCGUCUGUGAAUAGCUUUGAAUGAAGCAGCAUAACACGGCAGAUUAAAUAUAAUCGAAGGAACAUAGGUAGUGAUAAAGCUACGUCGUAUGGCACCAUUUCGGUGCCAAUUGUUUUAU ....((.((((....(((((....)))))..))))))(((((.(((((....)))))............(((((......)))))))))).(((((((.....))))))).......... ( -33.60) >DroWil_CAF1 6633 120 - 1 AUGCGUCUGUGAAUAGCUUUGAAUGUAACAGCAUUACGCGGCAGAUUAAAUAUAAUCGAAGGAACAUAGGUAAUGAUAAAGCUACGUCAUAUGGCACCAGUUCGGUGCCAAUUGUUUUGU .......(((((.((((((((..(((....((.....)).))).((((..(((..((....))..)))..))))..))))))))..)))))(((((((.....))))))).......... ( -31.40) >DroMoj_CAF1 9630 120 - 1 AUGCGUCUGUGAAUAGCUUUGAAUGUAGCAGCAUGACACGGCAGAUUAAAUAUAAUCGAAGGAACAUAGGUAGUGAUAAAGCUACGUCGUAGGGCACCAUUUCGGUGCCAAUUGUUUUAU .((((.(((((....(((.((.((((....))))..)).))).(((((....))))).......)))))(((((......)))))..)))).((((((.....))))))........... ( -33.60) >DroAna_CAF1 6575 120 - 1 AUGCGUCUGUGAAUAGUUUCGAAUGCAGCAGCAUUACGCGGCAGAUUAAAUAUAAUCGAAGGAACAUAGGUAAUGAUAAAGCUACGUCAUAUGGCACCAAUUCGGUGCCAAUUGUUUUAU .....((((((....((..(((((((....))))).))..)).(((((....))))).......))))))..(((((........))))).(((((((.....))))))).......... ( -31.50) >consensus AUGCGUCUGUGAAUAGCUUUGAAUGAAGCAGCAUUACGCGGCAGAUUAAAUAUAAUCGAAGGAACAUAGGUAAUGAUAAAGCUACGUCAUAUGGCACCAUUUCGGUGCCAAUUGUUUUAU .......(((((.((((((((.........((........))..((((..(((..((....))..)))..))))..))))))))..)))))(((((((.....))))))).......... (-26.19 = -26.08 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:01 2006