
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,220,662 – 5,220,790 |
| Length | 128 |
| Max. P | 0.981818 |

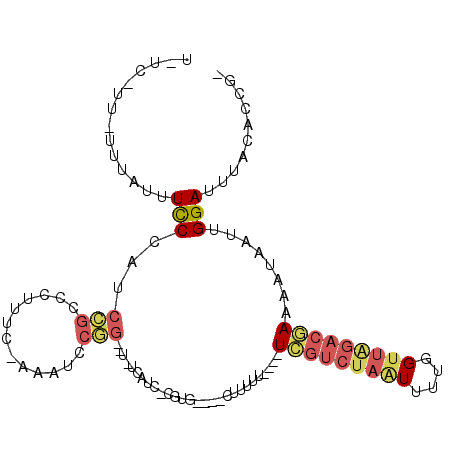
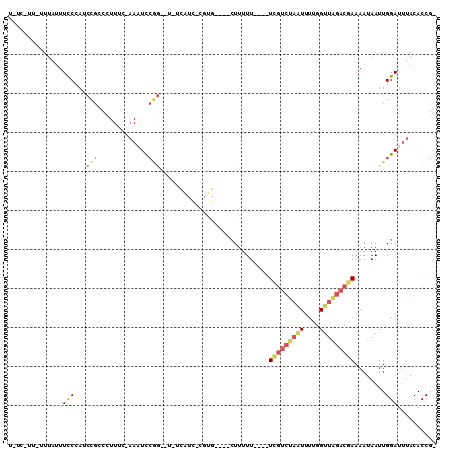
| Location | 5,220,662 – 5,220,759 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 71.80 |
| Mean single sequence MFE | -18.62 |
| Consensus MFE | -5.83 |
| Energy contribution | -6.74 |
| Covariance contribution | 0.91 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.31 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
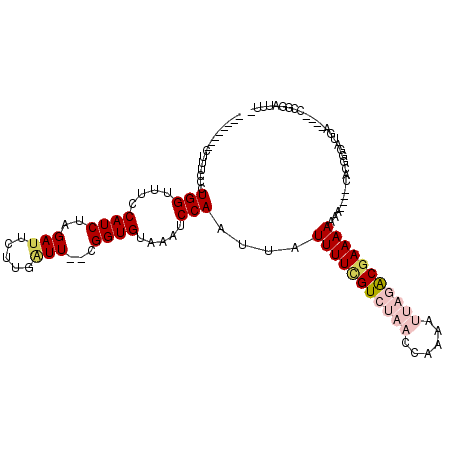
>X_DroMel_CAF1 5220662 97 + 22224390 U-UC-UU-UUUAUUUCCCAUCCGCCCUUUUGAAAUCCGGG-UAUCAUCUCGUG----CUUUUU----UCGUCUAAUUUUGGUUAGACGAAAAUAAUUGGAUUUACACCG- .-..-..-.....................(((((((((((-(((......)))----)))(((----(((((((((....)))))))))))).....)))))).))...- ( -21.80) >DroPse_CAF1 45637 93 + 1 U-UUGCU-GUUAUUUUCUGU-UUCGUUUAC-CAUUU-------UUGUUCCUUGAAUUUUUUUU----UCGC--UGUUUUGGUUAGACCAAAAUAAUUGGAUUUACACCCA .-.....-............-........(-((...-------.........(((.......)----))..--((((((((.....))))))))..)))........... ( -10.10) >DroSec_CAF1 40487 95 + 1 U-UC-UU-UU--UUUCCCAUCCGCCCUUUC-AAAUCCGGGCUAUCAUCUCGUG----UUUUUU----UCGUCUAAUUUUGGUUAGACGAAAAUAAUUGGAUGUACACCG- .-..-..-..--.....(((((((((....-......))))...........(----((.(((----(((((((((....)))))))))))).))).))))).......- ( -26.70) >DroEre_CAF1 44025 97 + 1 UUUC-UU-UUUAUUUCCCAACCGCCCUUUC-AAAUCCGGGCU-ACAUCCCGUG----CUUUUU----UCGUCUAAUUUUGGUUAGACGAAAAUAAUUGGAUUUACACCG- ....-..-..............((((....-......)))).-..((((.((.----...(((----(((((((((....))))))))))))..)).))))........- ( -23.60) >DroYak_CAF1 41311 83 + 1 U-UC-UU-UUUAUUUCCCGUCCGCUCUUUC-AAAUCCGG------------------CUUUUU----UCGUCUAAUUUUGGUUAGACGAAAAUAAUUGGAUUUACACCG- .-..-..-......................-((((((((------------------...(((----(((((((((....))))))))))))...))))))))......- ( -21.10) >DroAna_CAF1 47164 93 + 1 U-UC-CUCCUUUAUUCCGAGCUGCCCUUUC-AAUUCCAC--UUCCA-----------CUUUUUAUCCUUUUCUAUUUUUGGUUGGACAAAAAUAAUUGGAUUUACACCG- .-..-.........................-........--.....-----------......((((.....((((((((......))))))))...))))........- ( -8.40) >consensus U_UC_UU_UUUAUUUCCCAUCCGCCCUUUC_AAAUCCGG__U_UCAUC_CGUG____CUUUUU____UCGUCUAAUUUUGGUUAGACGAAAAUAAUUGGAUUUACACCG_ ..............(((...(((.............)))............................(((((((((....)))))))))........))).......... ( -5.83 = -6.74 + 0.91)

| Location | 5,220,662 – 5,220,759 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 71.80 |
| Mean single sequence MFE | -19.36 |
| Consensus MFE | -4.84 |
| Energy contribution | -6.30 |
| Covariance contribution | 1.46 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.25 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5220662 97 - 22224390 -CGGUGUAAAUCCAAUUAUUUUCGUCUAACCAAAAUUAGACGA----AAAAAG----CACGAGAUGAUA-CCCGGAUUUCAAAAGGGCGGAUGGGAAAUAAA-AA-GA-A -........((((.....(((((((((((......))))))))----)))...----............-(((...........))).))))..........-..-..-. ( -19.50) >DroPse_CAF1 45637 93 - 1 UGGGUGUAAAUCCAAUUAUUUUGGUCUAACCAAAACA--GCGA----AAAAAAAAUUCAAGGAACAA-------AAAUG-GUAAACGAA-ACAGAAAAUAAC-AGCAA-A (((((....)))))....((((((.....))))))..--((((----(.......))).........-------...((-.....))..-............-.))..-. ( -10.20) >DroSec_CAF1 40487 95 - 1 -CGGUGUACAUCCAAUUAUUUUCGUCUAACCAAAAUUAGACGA----AAAAAA----CACGAGAUGAUAGCCCGGAUUU-GAAAGGGCGGAUGGGAAA--AA-AA-GA-A -.......(((((.....(((((((((((......))))))))----)))...----............((((......-....))))))))).....--..-..-..-. ( -27.40) >DroEre_CAF1 44025 97 - 1 -CGGUGUAAAUCCAAUUAUUUUCGUCUAACCAAAAUUAGACGA----AAAAAG----CACGGGAUGU-AGCCCGGAUUU-GAAAGGGCGGUUGGGAAAUAAA-AA-GAAA -.........(((((((.(((((((((((......))))))))----)))...----..........-.((((......-....))))))))))).......-..-.... ( -25.50) >DroYak_CAF1 41311 83 - 1 -CGGUGUAAAUCCAAUUAUUUUCGUCUAACCAAAAUUAGACGA----AAAAAG------------------CCGGAUUU-GAAAGAGCGGACGGGAAAUAAA-AA-GA-A -(.((.(((((((..((.(((((((((((......))))))))----))).))------------------..))))))-).....)).)............-..-..-. ( -19.30) >DroAna_CAF1 47164 93 - 1 -CGGUGUAAAUCCAAUUAUUUUUGUCCAACCAAAAAUAGAAAAGGAUAAAAAG-----------UGGAA--GUGGAAUU-GAAAGGGCAGCUCGGAAUAAAGGAG-GA-A -((((.((..((((....(((((((((................))))))))).-----------)))).--.))..)))-).........(((.........)))-..-. ( -14.29) >consensus _CGGUGUAAAUCCAAUUAUUUUCGUCUAACCAAAAUUAGACGA____AAAAAG____CACG_GAUGA_A__CCGGAUUU_GAAAGGGCGGAUGGGAAAUAAA_AA_GA_A ..........(((........((((((((......))))))))............................(((.............)))...))).............. ( -4.84 = -6.30 + 1.46)

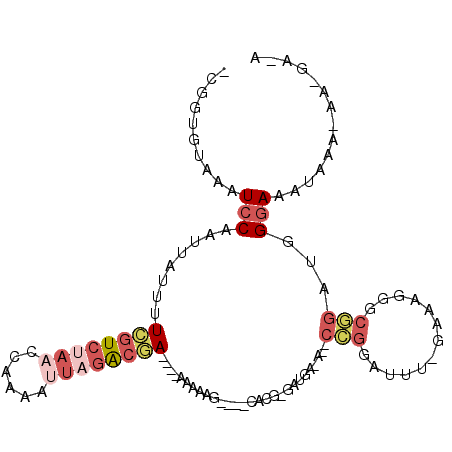
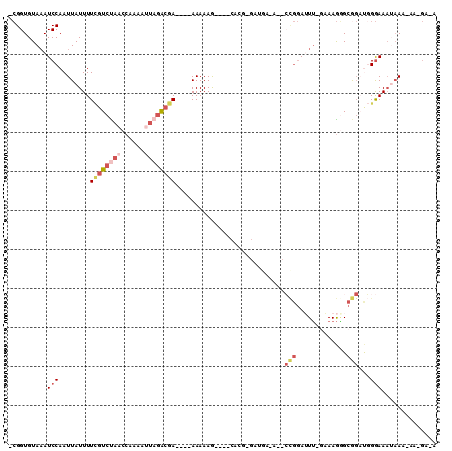
| Location | 5,220,689 – 5,220,790 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 73.49 |
| Mean single sequence MFE | -23.05 |
| Consensus MFE | -10.31 |
| Energy contribution | -11.48 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5220689 101 - 22224390 --------CUUUGCCUGGUUUCCAUCUAGAUUCUUGAUU--CGGUGUAAAUCCAAUUAUUUUCGUCUAACCAAAAUUAGACGAAAAAAG----CACGAGAUGAUA-CCCGGAUUUC --------.....((.(((...(((((.......(((((--.(((....))).)))))(((((((((((......)))))))))))...----....)))))..)-)).))..... ( -25.00) >DroPse_CAF1 45664 105 - 1 UGUGUGGACUUUCUCUGGUUU-CAUCUUGAUUCUUGGUUUUGGGUGUAAAUCCAAUUAUUUUGGUCUAACCAAAACA--GCGAAAAAAAAAUUCAAGGAACAA-------AAAUG- .(((.(((((......)))))-))).(((.(((((((..((((((....))))))...((((((.....))))))..--.............))))))).)))-------.....- ( -19.90) >DroSec_CAF1 40512 101 - 1 --------CUUUGCCUGGUUUCCAUCUAGAUUCUUGAUU--CGGUGUACAUCCAAUUAUUUUCGUCUAACCAAAAUUAGACGAAAAAAA----CACGAGAUGAUAGCCCGGAUUU- --------.....((.((((..(((((.......(((((--.(((....))).)))))(((((((((((......)))))))))))...----....)))))..)))).))....- ( -27.00) >DroEre_CAF1 44053 100 - 1 --------CUUUCCCUGGCCUCCAUCUAGACUCUUGAUU--CGGUGUAAAUCCAAUUAUUUUCGUCUAACCAAAAUUAGACGAAAAAAG----CACGGGAUGU-AGCCCGGAUUU- --------......((((.((.(((((.......(((((--.(((....))).)))))(((((((((((......)))))))))))...----....))))).-)).))))....- ( -26.40) >DroYak_CAF1 41338 87 - 1 --------CUUACCCUGGCCUCCAUCUAGAUUCUUGAUU--CGGUGUAAAUCCAAUUAUUUUCGUCUAACCAAAAUUAGACGAAAAAAG------------------CCGGAUUU- --------......(((((...............(((((--.(((....))).)))))(((((((((((......)))))))))))..)------------------))))....- ( -21.90) >DroPer_CAF1 46180 104 - 1 UGUGUGGACUUUCUCUGGUUU-CAUCUUGAUUCUUGGUUUUGGGUGUAAAUCCAAUUAUUUUUGUCGAACCAAAACA--GCGAAAA-AAAAUUCAAGGAACAA-------AAAUG- .(((.(((((......)))))-))).(((.(((((((..((((((....))))))...(((((((.(........).--)))))))-.....))))))).)))-------.....- ( -18.10) >consensus ________CUUUCCCUGGUUUCCAUCUAGAUUCUUGAUU__CGGUGUAAAUCCAAUUAUUUUCGUCUAACCAAAAUUAGACGAAAAAAA____CACGGGAUGA____CCGGAUUU_ ...............(((....((((..(((.....)))...)))).....)))....(((((((((((......))))))))))).............................. (-10.31 = -11.48 + 1.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:56 2006