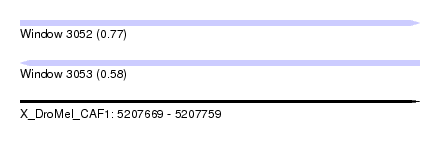
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,207,669 – 5,207,759 |
| Length | 90 |
| Max. P | 0.765214 |

| Location | 5,207,669 – 5,207,759 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 82.72 |
| Mean single sequence MFE | -23.96 |
| Consensus MFE | -17.57 |
| Energy contribution | -17.41 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
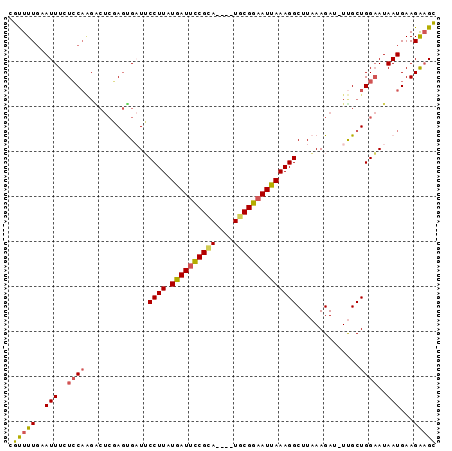
>X_DroMel_CAF1 5207669 90 + 22224390 CGUUUUGAAUUUCUCCAAGGCUCGAGUGUUUCCUUAUAAUUCCGCA----UGCGGAAUUAAAGGCUUAAAGAU-UUGCUGGAAUAAUGAAGAAGC .(((((..(((..((((..((..((((.(((((((.((((((((..----..))))))))))))...))).))-))))))))..)))...))))) ( -23.20) >DroSec_CAF1 28105 90 + 1 CGCUUUGAAUUUCUCCAAGACUCGAGUGAUUCCUUAUAAUUCCGCA----UGCGGAAUUAAAGGCUUAAAGAU-UUGCUGGAAUAAUGAAGAAGC .(((((..(((..((((.((.((...(((..((((.((((((((..----..)))))))))))).)))..)).-))..))))..)))...))))) ( -25.40) >DroSim_CAF1 21800 90 + 1 CGCUUUGAAUUUCUCCAAGACUCGAGUGAUUCCUUAUGAUUCCGCA----UGCGGAAUUAAAGGCUUAAAGAU-UUGCUGGAAUAAUGAAGAAGC .(((((..(((..((((.((.((...(((..((((.((((((((..----..)))))))))))).)))..)).-))..))))..)))...))))) ( -25.50) >DroEre_CAF1 31309 91 + 1 GGUUUUGAAUUUCUGCAGGGCUCUAGCGAUGCCUUAUGAUGCCGUA----UACGGAAUUAAAGGCUUUAAGACUUCGCUGGCAUAAUGAAGGAGU ........((((((.((.....(((((((.(((((.((((.(((..----..))).))))))))).........))))))).....)).)))))) ( -25.30) >DroYak_CAF1 28711 86 + 1 CGUUUUCAAUUUCUCC--------AUCGCUGCCUUAUGAUUUCGCAUGCGUACGGAAUUAAAGGCUUAAAGAU-UUGCUGGAAUAAUGAAGGUGU ((..((((.((..(((--------(((...(((((.((((((((........))))))))))))).....)).-....))))..))))))..)). ( -20.40) >consensus CGUUUUGAAUUUCUCCAAGACUCGAGUGAUUCCUUAUGAUUCCGCA____UGCGGAAUUAAAGGCUUAAAGAU_UUGCUGGAAUAAUGAAGAAGC .(((((..(((..((((..............((((.((((((((((....))))))))))))))..............))))..)))...))))) (-17.57 = -17.41 + -0.16)

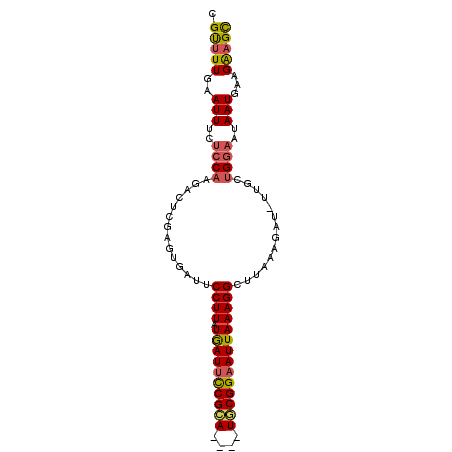
| Location | 5,207,669 – 5,207,759 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 82.72 |
| Mean single sequence MFE | -19.05 |
| Consensus MFE | -14.67 |
| Energy contribution | -15.35 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
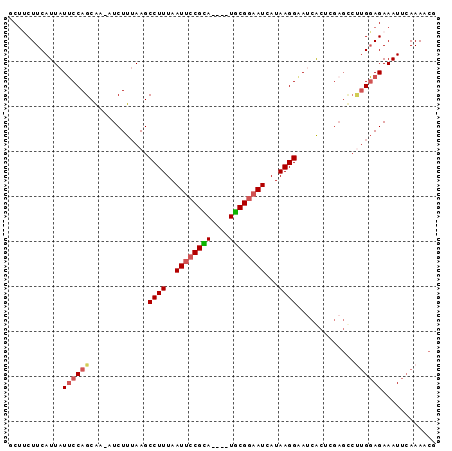
>X_DroMel_CAF1 5207669 90 - 22224390 GCUUCUUCAUUAUUCCAGCAA-AUCUUUAAGCCUUUAAUUCCGCA----UGCGGAAUUAUAAGGAAACACUCGAGCCUUGGAGAAAUUCAAAACG (((.............)))..-.((((((((.(((((((((((..----..))))))))...(....)....))).))))))))........... ( -20.82) >DroSec_CAF1 28105 90 - 1 GCUUCUUCAUUAUUCCAGCAA-AUCUUUAAGCCUUUAAUUCCGCA----UGCGGAAUUAUAAGGAAUCACUCGAGUCUUGGAGAAAUUCAAAGCG (((((((((..((((.((...-.((((((......((((((((..----..))))))))))))))....)).))))..)))))).......))). ( -23.01) >DroSim_CAF1 21800 90 - 1 GCUUCUUCAUUAUUCCAGCAA-AUCUUUAAGCCUUUAAUUCCGCA----UGCGGAAUCAUAAGGAAUCACUCGAGUCUUGGAGAAAUUCAAAGCG (((((((((..((((.((...-.........((((..((((((..----..))))))...)))).....)).))))..)))))).......))). ( -20.24) >DroEre_CAF1 31309 91 - 1 ACUCCUUCAUUAUGCCAGCGAAGUCUUAAAGCCUUUAAUUCCGUA----UACGGCAUCAUAAGGCAUCGCUAGAGCCCUGCAGAAAUUCAAAACC .............((((((((.((((((..(((............----...)))....)))))).))))).).))................... ( -16.96) >DroYak_CAF1 28711 86 - 1 ACACCUUCAUUAUUCCAGCAA-AUCUUUAAGCCUUUAAUUCCGUACGCAUGCGAAAUCAUAAGGCAGCGAU--------GGAGAAAUUGAAAACG .....((((...(((((((((-....))..(((((..(((.((((....)))).)))...))))).))..)--------))))....)))).... ( -14.20) >consensus GCUUCUUCAUUAUUCCAGCAA_AUCUUUAAGCCUUUAAUUCCGCA____UGCGGAAUCAUAAGGAAUCACUCGAGCCUUGGAGAAAUUCAAAACG ............((((((.............((((..((((((((....))))))))...)))).............))))))............ (-14.67 = -15.35 + 0.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:52 2006