
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,193,590 – 5,193,688 |
| Length | 98 |
| Max. P | 0.669796 |

| Location | 5,193,590 – 5,193,688 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.87 |
| Mean single sequence MFE | -26.39 |
| Consensus MFE | -14.88 |
| Energy contribution | -14.61 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
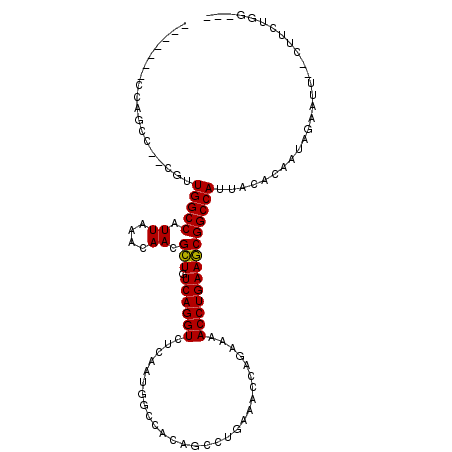
>X_DroMel_CAF1 5193590 98 + 22224390 -------CCAGCC--CGUUGGCCAUUAAACAACGCUGUCAGGUCUCAAUGGCCACAGCCUGAAACCAGAAAACCUGAAGCGGCCAAUAUACAAUAGAAUU--CUGCUGG--- -------(((((.--.(((((((..........(((((..((((.....)))))))))(((....)))............))))))).......((....--)))))))--- ( -31.20) >DroSec_CAF1 13548 98 + 1 -------CCAGCC--CGUUGGCCAUUAAACAACGCUGUCAGGUCUCAAUGACCACAGCCUGAAACCAGAAAACCUGAAGCGGCCAUUACACAAUAGAAUU--CUUCUGG--- -------((((..--.(((((((..........(((((..((((.....)))))))))(((....)))............)))))..)).....((....--)).))))--- ( -25.10) >DroSim_CAF1 7541 98 + 1 -------CCAGCC--CGUUGGCCAUUAAACAACGCUGUCAGGUCUCAAUGGCCACAGCCUGAAACCAGAAAACCUGAAGCGGCCAUUACACAAUAGAAUU--CUUCUGG--- -------((((..--.(((((((..........(((((..((((.....)))))))))(((....)))............)))))..)).....((....--)).))))--- ( -24.50) >DroWil_CAF1 25023 99 + 1 ----------GGUGGAGGUGGCCAUUAAACAACGGUGUCAGGUUUGAAUGGGCAAGGC---AAAAGAAAAAACCUGAAUCGGCCAUUAAACAAUAGAAUUUUCUUUCCUUUU ----------...((((((((((.....((....)).((((((((......((...))---........))))))))...))))))).......(((....))).))).... ( -21.94) >DroYak_CAF1 13821 105 + 1 GAACAGAACAGCC--CGUUGGCCAUUAAACAACGCUGUCAGGUCUCAAUGGCCACAGCCUGAAACCAGAAAACCUGAAGCGGCCAUUACACAAUAGAAUU--CUUCUGG--- ...(((((.....--.(((((((..........(((((..((((.....)))))))))(((....)))............)))))..))......(....--)))))).--- ( -25.40) >DroAna_CAF1 13538 101 + 1 AAGAAGGACCUGC--UGCUGGCCAUUAAACAACGCUGUCAGGUUUCAAUGGGCAGGACCUGC------AGAACCUGAAGCGGCCAUUACACAAUAGAAUUUCCUUCUCU--- .(((((((.(((.--((.(((((.((....)).(((.((((((((....((......))...------.)))))))))))))))).....)).)))....)))))))..--- ( -30.20) >consensus _______CCAGCC__CGUUGGCCAUUAAACAACGCUGUCAGGUCUCAAUGGCCACAGCCUGAAACCAGAAAACCUGAAGCGGCCAUUACACAAUAGAAUU__CUUCUGG___ ..................(((((.((....)).(((.((((((............................))))))))))))))........................... (-14.88 = -14.61 + -0.28)


| Location | 5,193,590 – 5,193,688 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.87 |
| Mean single sequence MFE | -30.96 |
| Consensus MFE | -18.89 |
| Energy contribution | -19.45 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5193590 98 - 22224390 ---CCAGCAG--AAUUCUAUUGUAUAUUGGCCGCUUCAGGUUUUCUGGUUUCAGGCUGUGGCCAUUGAGACCUGACAGCGUUGUUUAAUGGCCAACG--GGCUGG------- ---(((((((--....))........((((((((((((((((((.((((..(.....)..))))..))))))))).)))(((....)))))))))..--.)))))------- ( -35.10) >DroSec_CAF1 13548 98 - 1 ---CCAGAAG--AAUUCUAUUGUGUAAUGGCCGCUUCAGGUUUUCUGGUUUCAGGCUGUGGUCAUUGAGACCUGACAGCGUUGUUUAAUGGCCAACG--GGCUGG------- ---(((((((--(...(((((....)))))((......))))))))))...(((.((((((((((((((...((....))...)))))))))).)))--).))).------- ( -31.10) >DroSim_CAF1 7541 98 - 1 ---CCAGAAG--AAUUCUAUUGUGUAAUGGCCGCUUCAGGUUUUCUGGUUUCAGGCUGUGGCCAUUGAGACCUGACAGCGUUGUUUAAUGGCCAACG--GGCUGG------- ---(((((((--(...(((((....)))))((......))))))))))...(((.((((((((((((((...((....))...)))))))))).)))--).))).------- ( -33.80) >DroWil_CAF1 25023 99 - 1 AAAAGGAAAGAAAAUUCUAUUGUUUAAUGGCCGAUUCAGGUUUUUUCUUUU---GCCUUGCCCAUUCAAACCUGACACCGUUGUUUAAUGGCCACCUCCACC---------- ....(((.........((((((..((((((.....((((((((........---((...))......))))))))..))))))..)))))).....)))...---------- ( -18.18) >DroYak_CAF1 13821 105 - 1 ---CCAGAAG--AAUUCUAUUGUGUAAUGGCCGCUUCAGGUUUUCUGGUUUCAGGCUGUGGCCAUUGAGACCUGACAGCGUUGUUUAAUGGCCAACG--GGCUGUUCUGUUC ---(((((((--(...(((((....)))))((......))))))))))...(((.((((((((((((((...((....))...)))))))))).)))--).)))........ ( -33.10) >DroAna_CAF1 13538 101 - 1 ---AGAGAAGGAAAUUCUAUUGUGUAAUGGCCGCUUCAGGUUCU------GCAGGUCCUGCCCAUUGAAACCUGACAGCGUUGUUUAAUGGCCAGCA--GCAGGUCCUUCUU ---.((((((((.......((((....((((((((((((((((.------((((...)))).....).))))))).)))(((....))))))))...--)))).)))))))) ( -34.50) >consensus ___CCAGAAG__AAUUCUAUUGUGUAAUGGCCGCUUCAGGUUUUCUGGUUUCAGGCUGUGGCCAUUGAGACCUGACAGCGUUGUUUAAUGGCCAACG__GGCUGG_______ ...........................(((((((((((((((((......(((.....))).....))))))))).)))(((....)))))))).................. (-18.89 = -19.45 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:45 2006