
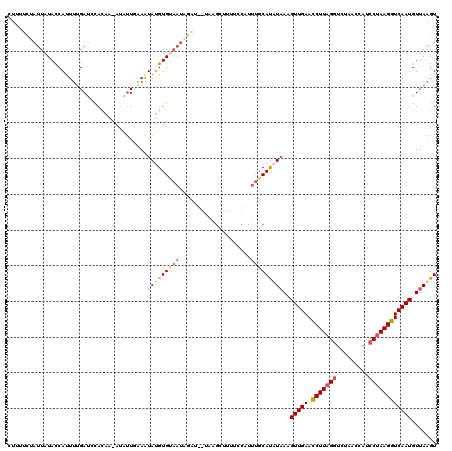
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,183,097 – 5,183,275 |
| Length | 178 |
| Max. P | 0.990351 |

| Location | 5,183,097 – 5,183,202 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.36 |
| Mean single sequence MFE | -22.60 |
| Consensus MFE | -12.49 |
| Energy contribution | -13.92 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5183097 105 + 22224390 CUUUUCUAUUAUACCACUUUGAUCCACUAUAUAUUUUAAUAUGUGCAAUAGAU---------------UUGCAUUUAAGGUUGAAUCUUAGGUCUAACUAUCCUAAGGUCAAUGUUAAGU ............................((((((....))))))((((.....---------------))))((((((.(((((.(((((((.........)))))))))))).)))))) ( -20.60) >DroSec_CAF1 4398 113 + 1 -------AUUAUACCAUUUUGAUUCACAAUAUGUUUAAAUAUGUGUAAUAGAUUGUUAGCUUUUGCAUUUGCAUUUAAGGUUGAAUCUUAGGUCUAACUAUCCUAAGGUCAAUGUUAAGU -------(((((((...(((((..((.....)).)))))...))))))).........((....))......((((((.(((((.(((((((.........)))))))))))).)))))) ( -22.50) >DroEre_CAF1 4379 108 + 1 CUGUUCUAUUAUAUCAUUUUGAUCCAUAA--GAUUGAAGUAUGUG----------UAAGCUUUUCCAUUUGCACAUUAAGUUGAACCUUCGCUCUUACCAUCCUAAGGUCAAUGUUAAGU ..((((.(((.....((((..(((.....--)))..))))(((((----------((((........)))))))))..))).))))(((.((....(((.......)))....)).))). ( -18.60) >DroYak_CAF1 4303 119 + 1 CUUAUCUAUUAUACCAUUGGGAGCUAUAA-ACAAUGCAAUAUGUGUAAUAGAUUGUAAGCUUUUCCAUUUGCAUAUAAAGUUGAACCUUAGGUCUAACCAUCCUAAGGUCAAUGUUAAGU (((((((((((((((((((..........-.)))))......))))))))))(((((.((..........)).))))).(((((.(((((((.........))))))))))))..)))). ( -28.70) >consensus CUUUUCUAUUAUACCAUUUUGAUCCACAA_AUAUUGAAAUAUGUGUAAUAGAU__UAAGCUUUUCCAUUUGCAUAUAAAGUUGAACCUUAGGUCUAACCAUCCUAAGGUCAAUGUUAAGU .......................................(((((((((....................)))))))))..(((((.(((((((.........))))))))))))....... (-12.49 = -13.92 + 1.44)


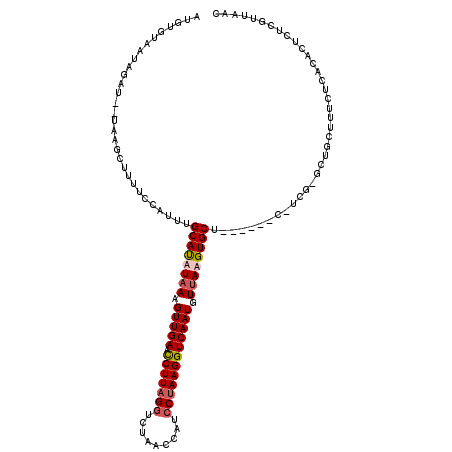
| Location | 5,183,137 – 5,183,235 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.02 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -19.19 |
| Energy contribution | -20.00 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5183137 98 + 22224390 AUGUGCAAUAGAU---------------UUGCAUUUAAGGUUGAAUCUUAGGUCUAACUAUCCUAAGGUCAAUGUUAAGUGCU------U-UCGGGCUGCUUUCUCACACUCUCGUUAAC ....(((...((.---------------..((((((((.(((((.(((((((.........)))))))))))).)))))))).------.-))....))).................... ( -26.90) >DroSec_CAF1 4431 112 + 1 AUGUGUAAUAGAUUGUUAGCUUUUGCAUUUGCAUUUAAGGUUGAAUCUUAGGUCUAACUAUCCUAAGGUCAAUGUUAAGUGCU------C-UCG-GCUGCUUUCUCGCACUCUCGUUAAC ..((((...(((..((.((((.........((((((((.(((((.(((((((.........)))))))))))).)))))))).------.-..)-)))))..))).)))).......... ( -32.02) >DroEre_CAF1 4417 103 + 1 AUGUG----------UAAGCUUUUCCAUUUGCACAUUAAGUUGAACCUUCGCUCUUACCAUCCUAAGGUCAAUGUUAAGUGCU------CCUCG-GCUGCUUUCUCAUACUCUCGUUAAC .((((----------.((((....((....((((.(((((((((.((((...............))))))))).)))))))).------....)-)..))))...))))........... ( -18.46) >DroYak_CAF1 4342 119 + 1 AUGUGUAAUAGAUUGUAAGCUUUUCCAUUUGCAUAUAAAGUUGAACCUUAGGUCUAACCAUCCUAAGGUCAAUGUUAAGUGCUUUCUCUGCUCG-UCUGCUUUCUCCUACUAUCGUUAAC ....(((..(((..(((..(..........((((.(((.(((((.(((((((.........)))))))))))).))).))))...........)-..)))..)))..))).......... ( -27.40) >consensus AUGUGUAAUAGAU__UAAGCUUUUCCAUUUGCAUAUAAAGUUGAACCUUAGGUCUAACCAUCCUAAGGUCAAUGUUAAGUGCU______C_UCG_GCUGCUUUCUCACACUCUCGUUAAC ..............................((((((((.(((((.(((((((.........)))))))))))).))))))))...................................... (-19.19 = -20.00 + 0.81)


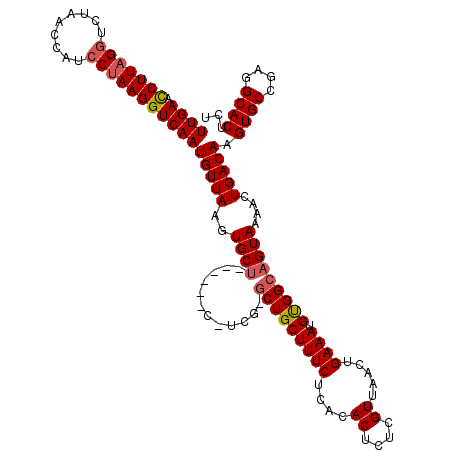
| Location | 5,183,162 – 5,183,275 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.40 |
| Mean single sequence MFE | -31.59 |
| Consensus MFE | -23.31 |
| Energy contribution | -23.88 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5183162 113 + 22224390 UUGAAUCUUAGGUCUAACUAUCCUAAGGUCAAUGUUAAGUGCU------U-UCGGGCUGCUUUCUCACACUCUCGUUAACUGAAAUGUGGCAGUAAAACUGACAAGUGCCGAGGCACUCU ((((.(((((((.........))))))))))).....((((((------(-.((((((((.....((((...(((.....)))..)))))))))...(((....))).)))))))))).. ( -31.50) >DroSec_CAF1 4471 112 + 1 UUGAAUCUUAGGUCUAACUAUCCUAAGGUCAAUGUUAAGUGCU------C-UCG-GCUGCUUUCUCGCACUCUCGUUAACUGAAAUGUGGCAGUAAAACUGACAAGUGCCGAGGCACUCU ((((.(((((((.........))))))))))).....(((((.------(-(((-((.((((..(((((...(((.....)))..)))))(((.....)))..))))))))))))))).. ( -34.80) >DroEre_CAF1 4447 113 + 1 UUGAACCUUCGCUCUUACCAUCCUAAGGUCAAUGUUAAGUGCU------CCUCG-GCUGCUUUCUCAUACUCUCGUUAACUGAAAUGCGGCCGUAAAACUGACAAGUGCCGAGGCACUCU ((((.((((...............))))))))(((((.((...------...((-(((((((((....((....)).....)))).)))))))....)))))))(((((....))))).. ( -29.86) >DroYak_CAF1 4382 119 + 1 UUGAACCUUAGGUCUAACCAUCCUAAGGUCAAUGUUAAGUGCUUUCUCUGCUCG-UCUGCUUUCUCCUACUAUCGUUAACUGAAAUGUGGCAGUAAAACUGACAAGUGCCGGGGCACACU ((((.(((((((.........)))))))))))......(((((((...((((.(-((.(.(((((.((((..(((.....)))...)))).)).))).).))).))))..)))))))... ( -30.20) >consensus UUGAACCUUAGGUCUAACCAUCCUAAGGUCAAUGUUAAGUGCU______C_UCG_GCUGCUUUCUCACACUCUCGUUAACUGAAAUGUGGCAGUAAAACUGACAAGUGCCGAGGCACUCU ((((.(((((((.........)))))))))))(((((..((((............(((((((((....((....)).....)))).)))))))))....))))).((((....))))... (-23.31 = -23.88 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:43 2006