| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,145,601 – 5,145,700 |
| Length | 99 |
| Max. P | 0.820759 |

| Location | 5,145,601 – 5,145,700 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.19 |
| Mean single sequence MFE | -23.11 |
| Consensus MFE | -15.68 |
| Energy contribution | -16.68 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5145601 99 + 22224390 CCCUUUUUUAUUUCUGCCAGUGCACCCU-UGGCUUUUAAUGAGCUUCGAACGCGAGCCAAUGCACAUA--------------UAUAUAUUUUUAGCGCAGAAAUCAUCUGCAUA .........(((((((((.(((((...(-((((((.......((.......))))))))))))))...--------------..((......))).)))))))).......... ( -21.51) >DroSec_CAF1 26837 92 + 1 CCCUUUUU---UUCUGCCAGUGCAGCCC-UGGCAUUUAAUGAGCUUCGAACACGAGCCAAUGCACAUA------------------UAUUUUUAGCGCAGAAAUCAUCUGCAUA ........---........(((((((..-..))......((.(((.((....))))))).)))))...------------------..........(((((.....)))))... ( -21.20) >DroSim_CAF1 20691 92 + 1 CCCAUUUU---UUCUGCCAGUGCACCCU-UGGCUUUUAAUGAGCUUCGAACGCGAGCCAAUGCACAUA------------------UAUUUUUAGCGCAGAAAUCAUCUGCAUA ........---........(((((...(-((((((.......((.......))))))))))))))...------------------..........(((((.....)))))... ( -21.11) >DroYak_CAF1 23193 106 + 1 CCC--UUU---UUCUGCCAGUGCAGCCCGGGGCUUUUAAUGAGAAUCGAACUCGAGCAACUGCACACAUAUGUACAUAUGUAUAUAUAUUUUUAGCGCAGAAAU---CUGCGUA (((--(..---..((((....))))...)))).......(((((((((....)))((....))....((((((((....)))))))).))))))((((((....---)))))). ( -28.60) >consensus CCCUUUUU___UUCUGCCAGUGCACCCC_UGGCUUUUAAUGAGCUUCGAACGCGAGCCAAUGCACAUA__________________UAUUUUUAGCGCAGAAAUCAUCUGCAUA ...................(((((.....((((((....((.....)).....)))))).)))))...............................(((((.....)))))... (-15.68 = -16.68 + 1.00)


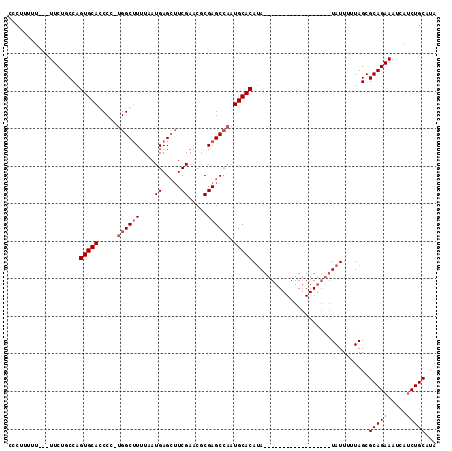
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:36 2006