| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,132,692 – 5,132,842 |
| Length | 150 |
| Max. P | 0.594592 |

| Location | 5,132,692 – 5,132,804 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -29.54 |
| Consensus MFE | -15.84 |
| Energy contribution | -16.72 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5132692 112 + 22224390 UUGUGCAGUUUAUCAAUCUCGAAUUUGAUUUCCUAGUGUGAAAUCUGACACGCGAUUCGCAUCAUUGCGAUUCGAUUGU-UGAGCACCAUGUGACAAAUUCGUCCAAUGGCAC ..((((.(((((.(((((.................(((((........)))))((.(((((....))))).))))))).-)))))..(((..(((......)))..))))))) ( -32.60) >DroVir_CAF1 17416 86 + 1 UCAAUGAGUUUAUCAAUUU-AAAUUUGAUUCGGUUGUGUGAAAAUCGACACGCGGCU--------------------AU-UUGGCGCUGCGUGACAAAUUUGCACAG-----C .....(((((((......)-))))))......((((((..((......((((((((.--------------------..-.....)))))))).....))..)))))-----) ( -25.80) >DroSec_CAF1 8915 112 + 1 UUGUGCAGUUUAUCAAUCUCGAAUUUGAUUUCCUAGUGUGAAAUCUGACACGCGAUUCGCAUCAUUGCGAUUCGAUUGU-UGGGCACCAUGUGACAAAUUCGUCCAAUGGCAC ..((((.(((((.(((((.................(((((........)))))((.(((((....))))).))))))).-)))))..(((..(((......)))..))))))) ( -31.70) >DroSim_CAF1 8684 112 + 1 UUGUGCAGUUUAUCAAUCUCGAAUUUGAUUUCCUAGUGUGAAAUCUGACACGCGAUUCGCAUCAUUGCGAUUCGAUUGU-UGGGCACCAUGUGACAAAUUCGUCCAAUGGCAC ..((((.(((((.(((((.................(((((........)))))((.(((((....))))).))))))).-)))))..(((..(((......)))..))))))) ( -31.70) >DroYak_CAF1 9814 113 + 1 UUGAGCAGUUUAUCAAUCUGGAAUUUGAUUUCCUAGUGUGAAAUCUGACACGCGAUCCGCAUCAUUGCAAUUCGAUUGUUUGGGGACUAUGUGACAAAUUCGUCCUAUGGCAC (..(((((((.(((.....((((......))))..(((((........))))))))..(((....))).....)))))))..).(.(((((.(((......))).)))))).. ( -25.90) >consensus UUGUGCAGUUUAUCAAUCUCGAAUUUGAUUUCCUAGUGUGAAAUCUGACACGCGAUUCGCAUCAUUGCGAUUCGAUUGU_UGGGCACCAUGUGACAAAUUCGUCCAAUGGCAC ....................(((((((........(((((........)))))((.(((((....))))).)).....................)))))))............ (-15.84 = -16.72 + 0.88)

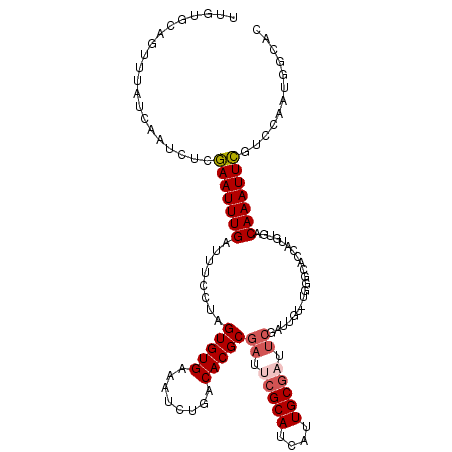

| Location | 5,132,692 – 5,132,804 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -26.89 |
| Consensus MFE | -14.74 |
| Energy contribution | -17.34 |
| Covariance contribution | 2.60 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.55 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5132692 112 - 22224390 GUGCCAUUGGACGAAUUUGUCACAUGGUGCUCA-ACAAUCGAAUCGCAAUGAUGCGAAUCGCGUGUCAGAUUUCACACUAGGAAAUCAAAUUCGAGAUUGAUAAACUGCACAA (..((((..((((....))))..))))..)(((-(...((((((.....(((((((.....)))))))((((((.......))))))..))))))..))))............ ( -30.70) >DroVir_CAF1 17416 86 - 1 G-----CUGUGCAAAUUUGUCACGCAGCGCCAA-AU--------------------AGCCGCGUGUCGAUUUUCACACAACCGAAUCAAAUUU-AAAUUGAUAAACUCAUUGA .-----.((((.(((.(((.(((((.((.....-..--------------------.)).))))).))).))))))).......(((((....-...)))))........... ( -20.20) >DroSec_CAF1 8915 112 - 1 GUGCCAUUGGACGAAUUUGUCACAUGGUGCCCA-ACAAUCGAAUCGCAAUGAUGCGAAUCGCGUGUCAGAUUUCACACUAGGAAAUCAAAUUCGAGAUUGAUAAACUGCACAA (..((((..((((....))))..))))..)...-.(((((((.(((((....))))).)).((.((..((((((.......))))))..)).)).)))))............. ( -29.60) >DroSim_CAF1 8684 112 - 1 GUGCCAUUGGACGAAUUUGUCACAUGGUGCCCA-ACAAUCGAAUCGCAAUGAUGCGAAUCGCGUGUCAGAUUUCACACUAGGAAAUCAAAUUCGAGAUUGAUAAACUGCACAA (..((((..((((....))))..))))..)...-.(((((((.(((((....))))).)).((.((..((((((.......))))))..)).)).)))))............. ( -29.60) >DroYak_CAF1 9814 113 - 1 GUGCCAUAGGACGAAUUUGUCACAUAGUCCCCAAACAAUCGAAUUGCAAUGAUGCGGAUCGCGUGUCAGAUUUCACACUAGGAAAUCAAAUUCCAGAUUGAUAAACUGCUCAA (.((....((((..............)))).....((((((((((....(((((((.....)))))))((((((.......)))))).)))))..))))).......)).).. ( -24.34) >consensus GUGCCAUUGGACGAAUUUGUCACAUGGUGCCCA_ACAAUCGAAUCGCAAUGAUGCGAAUCGCGUGUCAGAUUUCACACUAGGAAAUCAAAUUCGAGAUUGAUAAACUGCACAA (((((((..((((....))))..))))))).....(((((((.(((((....))))).))........((((((.......))))))........)))))............. (-14.74 = -17.34 + 2.60)

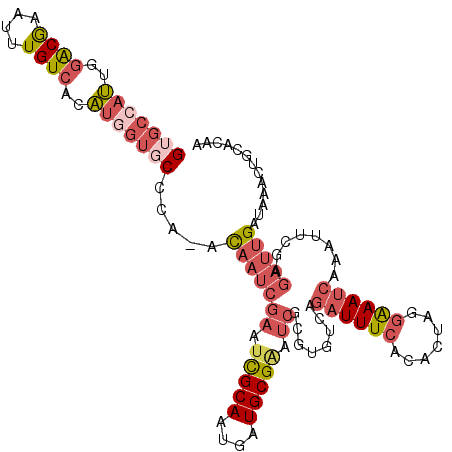

| Location | 5,132,727 – 5,132,842 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.22 |
| Mean single sequence MFE | -33.55 |
| Consensus MFE | -11.30 |
| Energy contribution | -12.86 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5132727 115 + 22224390 GUGUGAAAUCUGACACGCGAUUCGCAUCAUUGCGAUUCGAUUGU-UGAGCACCAUGUGACAAAUUCGUCCAAUGGCACGAAUAAUACCACCAUUUUUGC-UGGCAACAU-GCCACGCC (((((...((.((((..(((.(((((....))))).)))..)))-)))....(((((.....((((((((...)).))))))....(((.((....)).-)))..))))-).))))). ( -32.60) >DroVir_CAF1 17450 91 + 1 GUGUGAAAAUCGACACGCGGCU--------------------AU-UUGGCGCUGCGUGACAAAUUUGCACAG-----CCAGCGAGAUCGGAAUUCGGUCGGGGCGCCAC-GCGGUGCC (((..((......((((((((.--------------------..-.....)))))))).....))..)))..-----.......(((((.....)))))..((((((..-..)))))) ( -34.80) >DroSec_CAF1 8950 115 + 1 GUGUGAAAUCUGACACGCGAUUCGCAUCAUUGCGAUUCGAUUGU-UGGGCACCAUGUGACAAAUUCGUCCAAUGGCACGAAUAAUACCUCCAUUUUUGC-UGCCAACAU-GCUACGCC (((((...((..(((..(((.(((((....))))).)))..)))-..))...((((((((......)))...(((((((((.............)))).-)))))))))-).))))). ( -34.72) >DroSim_CAF1 8719 115 + 1 GUGUGAAAUCUGACACGCGAUUCGCAUCAUUGCGAUUCGAUUGU-UGGGCACCAUGUGACAAAUUCGUCCAAUGGCACGAAUAAUACCUCCAUUUUUGC-UGCCAACAU-GCCACGCC (((((...((..(((..(((.(((((....))))).)))..)))-..))...((((((((......)))...(((((((((.............)))).-)))))))))-).))))). ( -36.82) >DroYak_CAF1 9849 117 + 1 GUGUGAAAUCUGACACGCGAUCCGCAUCAUUGCAAUUCGAUUGUUUGGGGACUAUGUGACAAAUUCGUCCUAUGGCACGAAUAAUACCUCCAUUUCUGC-UGCCAACAUAGCCACGCC ((((....((.((((..(((...(((....)))...)))..)))).))((.(((((((((......)))...(((((((((............))).).-))))))))))))))))). ( -28.80) >consensus GUGUGAAAUCUGACACGCGAUUCGCAUCAUUGCGAUUCGAUUGU_UGGGCACCAUGUGACAAAUUCGUCCAAUGGCACGAAUAAUACCUCCAUUUUUGC_UGCCAACAU_GCCACGCC (((((........(((((((.(((((....))))).)))......((.....))))))....(((((((.....).))))))..............................))))). (-11.30 = -12.86 + 1.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:29 2006