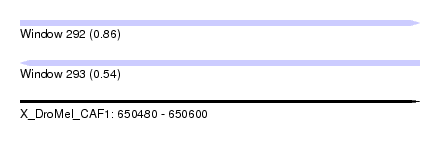
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 650,480 – 650,600 |
| Length | 120 |
| Max. P | 0.863459 |

| Location | 650,480 – 650,600 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -52.57 |
| Consensus MFE | -46.58 |
| Energy contribution | -46.92 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 650480 120 + 22224390 UCCCUCAUCAACAUGCACUUUGAGGGACGCUGGCUGGUCGCCAAGGAGCAGUUGCCCCUGCUGCUGGCGCAGAGCCUGCAACUGAUUCAGAAGCUGGUGACCAGGCAGCCCGACUACGCC (((((((.............))))))).((((.((((((((((..((((((((((.....((((....)))).....))))))).)))......)))))))))).))))........... ( -54.72) >DroEre_CAF1 8377 120 + 1 UCCCUCAUCAGCAUGCACUUUGAGGGCAGCUGGCUGGUCGCCAAGGAGCAGUUGCCACUGCUGCUGGCGCAGAGCCUGCAACUGAUUCAGAAGCUGGUGACCAAGCAGCCCGACUACGCG .((((((..((......)).))))))..((((..(((((((((..((((((((((.....((((....)))).....))))))).)))......)))))))))..))))........... ( -47.20) >DroYak_CAF1 8718 120 + 1 UCGCUCAUCAACAUGCACUUGGAGGGCUGCUGGCUGGUCGCCAAGGAGCAGUUGCCCCUGCUGCUGGCGCAGAGUCUGCAGUUGACUCAGAAGCUGGUGACCAGGCAGCCCGACUAUGCG ..............(((...(..(((((((...(((((((((.((.(((((......))))).))(((...(((((.......)))))....)))))))))))))))))))..)..))). ( -55.80) >consensus UCCCUCAUCAACAUGCACUUUGAGGGCAGCUGGCUGGUCGCCAAGGAGCAGUUGCCCCUGCUGCUGGCGCAGAGCCUGCAACUGAUUCAGAAGCUGGUGACCAGGCAGCCCGACUACGCG .((((((.............))))))..((((.((((((((((..((((((((((.....((((....)))).....))))))).)))......)))))))))).))))........... (-46.58 = -46.92 + 0.34)


| Location | 650,480 – 650,600 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -52.01 |
| Consensus MFE | -42.61 |
| Energy contribution | -42.07 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 650480 120 - 22224390 GGCGUAGUCGGGCUGCCUGGUCACCAGCUUCUGAAUCAGUUGCAGGCUCUGCGCCAGCAGCAGGGGCAACUGCUCCUUGGCGACCAGCCAGCGUCCCUCAAAGUGCAUGUUGAUGAGGGA ...........((((.((((((.(((............(((((.(((.....))).)))))((((((....))))))))).)))))).)))).(((((((.............))))))) ( -51.92) >DroEre_CAF1 8377 120 - 1 CGCGUAGUCGGGCUGCUUGGUCACCAGCUUCUGAAUCAGUUGCAGGCUCUGCGCCAGCAGCAGUGGCAACUGCUCCUUGGCGACCAGCCAGCUGCCCUCAAAGUGCAUGCUGAUGAGGGA ..........(((((.((((((.((((.....((..(((((((..(((((((....)))).))).))))))).)).)))).)))))).))))).((((((.(((....)))..)))))). ( -51.20) >DroYak_CAF1 8718 120 - 1 CGCAUAGUCGGGCUGCCUGGUCACCAGCUUCUGAGUCAACUGCAGACUCUGCGCCAGCAGCAGGGGCAACUGCUCCUUGGCGACCAGCCAGCAGCCCUCCAAGUGCAUGUUGAUGAGCGA .((((.(..(((((((((((((.(((((....(((((.......)))))(((....)))))((((((....))))))))).))))))...)))))))..)..)))).((((....)))). ( -52.90) >consensus CGCGUAGUCGGGCUGCCUGGUCACCAGCUUCUGAAUCAGUUGCAGGCUCUGCGCCAGCAGCAGGGGCAACUGCUCCUUGGCGACCAGCCAGCAGCCCUCAAAGUGCAUGUUGAUGAGGGA .(((.....(((((((((((((..(((..((((((....)).))))..))).(((((.(((((......))))).).))))))))))...)))))))......))).............. (-42.61 = -42.07 + -0.55)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:27 2006