
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,131,482 – 5,131,715 |
| Length | 233 |
| Max. P | 0.799094 |

| Location | 5,131,482 – 5,131,602 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -36.34 |
| Consensus MFE | -33.08 |
| Energy contribution | -32.45 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5131482 120 + 22224390 GUACUUGGCCCCCUUUUAUGGCCGAAAACGAAGAGGAAUUGUCAUGAAGCGGACAAACUUUUGUGCCACGAAUCCUUUGGGCGCAACUCCUUGUCGAUCCUUUUUCUGUUCCUGCAAAUG (((.((((((.........))))))...(((.(((((.(((((........)))))....(((((((.(((.....)))))))))).))))).)))................)))..... ( -32.50) >DroSec_CAF1 7711 120 + 1 GUACUUGGCCCCCUUUUAUGGCCGAAAACGAAAAGGAAUUGUCAUGAAGCGGACAAGCUUUUGCGCCACGAAUCCUUUGGGCGCAACUCCUUGUCGGUCCUUUUUCUGUUCCACCACAUG ((..((((((.........))))))..)).....(((((.........((.((((((...(((((((.(((.....))))))))))...)))))).)).........)))))........ ( -37.77) >DroSim_CAF1 7517 120 + 1 GUACUUGGCCCCCUUUUAUGGCCGAAAACGAAGAGGAAUUGUCAUGAAGCGGACAAACUUUUGCGCCACGAAUCCUUUGGGCGCAACUCCUUGUCGGUCCUUUUUCUGUUCCUCCAAAUG ((..((((((.........))))))..))...(((((((.........((.(((((....(((((((.(((.....))))))))))....))))).)).........)))))))...... ( -38.27) >DroYak_CAF1 8552 119 + 1 GUACUUGGCC-UCUUUUAUGGCCGAAAACGAAGAGGAAUUGUCAUGAAGCGGGCAAACUUUUGUGCCACGAAUCCUUUGGGCGCAACUCCUUGUCGGUCCUUUUUUUGGUUCUCCAAAUG ....((((((-........))))))...(((.(((((.(((((........)))))....(((((((.(((.....)))))))))).))))).)))........(((((....))))).. ( -36.80) >consensus GUACUUGGCCCCCUUUUAUGGCCGAAAACGAAGAGGAAUUGUCAUGAAGCGGACAAACUUUUGCGCCACGAAUCCUUUGGGCGCAACUCCUUGUCGGUCCUUUUUCUGUUCCUCCAAAUG ....((((((.........))))))...(((.(((((.(((((........)))))....(((((((.(((.....)))))))))).))))).)))........................ (-33.08 = -32.45 + -0.63)


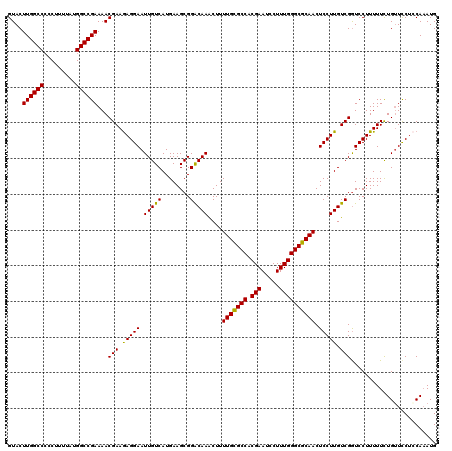
| Location | 5,131,602 – 5,131,715 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 85.80 |
| Mean single sequence MFE | -19.90 |
| Consensus MFE | -14.06 |
| Energy contribution | -14.06 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
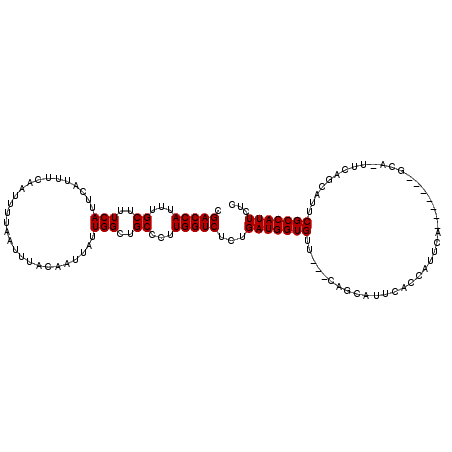
>X_DroMel_CAF1 5131602 113 - 22224390 CGACCAUUUGCUUUCAUUCAUUUCAAUUUUAAUUUACAAUUAUUGGCUGCCCUUGGUCUCCGAUGGUGGUGAACAGCAUUCAGCAUUUA------AGCAUUUCGCCAUUCGCCAUUCUC .(((((...((..(((...........................)))..))...)))))...((..((((((((..((.....((.....------.)).....))..))))))))..)) ( -23.83) >DroSec_CAF1 7831 108 - 1 CGACCAUUUGCUUUCAUUCAUUUCAAUUUUAAUUUACAAUUAUUGGCUGCCCUUGGUCUCUGAUGGUGUU---CAGCAUUCACCAUUCA-------CCA-UUCAGCAUUCGCCAUUCUC ...........................................(((((((...((((....(((((((..---.......))))))).)-------)))-....)))...))))..... ( -18.30) >DroSim_CAF1 7637 94 - 1 CGACCAUUUGCUUUCAUUCAUUUCAAUUUUAAUUUACAAUUAUUGGCUGCCCUUGGUCUCUGAUGGUGUU---CAGCAU----------------------UCAGCAUUCGCCAUUCUC .(((((...((..(((...........................)))..))...)))))...(((((((..---..((..----------------------...))...)))))))... ( -17.93) >DroYak_CAF1 8671 115 - 1 CGACCAUUUGCUUUCAUUCAUUUCAAUUUUAAUUUACAAUUAUUGGCUGCCCUUGGUCUCUGAUGGUGUU---CACCAUUCACCAUUCAGCAUUCAGCA-UUCAGCAUUCGCCAUUCUC .(((((...((..(((...........................)))..))...))))).(((((((((..---.......))))).))))......((.-....))............. ( -19.53) >consensus CGACCAUUUGCUUUCAUUCAUUUCAAUUUUAAUUUACAAUUAUUGGCUGCCCUUGGUCUCUGAUGGUGUU___CAGCAUUCACCAUUCA_______GCA_UUCAGCAUUCGCCAUUCUC .(((((...((..(((...........................)))..))...)))))...(((((((.........................................)))))))... (-14.06 = -14.06 + -0.00)

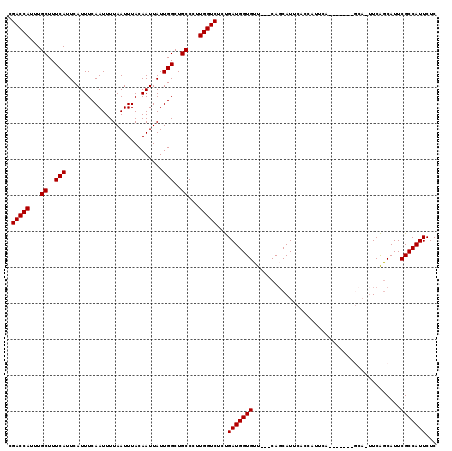
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:26 2006