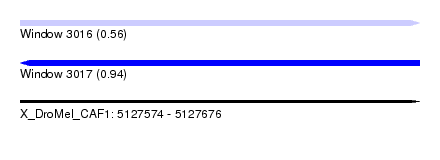
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,127,574 – 5,127,676 |
| Length | 102 |
| Max. P | 0.937571 |

| Location | 5,127,574 – 5,127,676 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 72.15 |
| Mean single sequence MFE | -31.23 |
| Consensus MFE | -20.24 |
| Energy contribution | -21.13 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5127574 102 + 22224390 GCUGAGGGUGAGG---GGUGGAAGAGGUCCAGGACAUGC--GACAGUUGGGAACACAGUGGGAGCGGCCGGCUGAAGCCAGCUAAAUUGCAAGUGGGUCACUGGGCC .............---.........((((((((((.(((--((.((((((.....((((.((.....)).))))...))))))...))))).....))).))))))) ( -33.60) >DroSec_CAF1 3967 99 + 1 ----NN-NNNNNN---NNNNNNNGGAGUCCAGGAGAUGCUGGGAUGUUGGAAAAACAGUGGGAGCGGCCGGCUGAAGCCAGCUAAAUUGCAAGUGGGUCACUGGGCC ----..-......---..........((((((((..(((((.((((((((.....((((.((.....)).))))...)))))...))).).))))..)).)))))). ( -28.90) >DroYak_CAF1 4636 105 + 1 GAGGAGGGGUAGGGGUAGGGGAUGGAGUCCAGAAGACGC--GACGGGGUGGAAAACACUGGGAGCGGCCGGCUGAAGCCAGCUAAAUUGCAAGUGGGUCAGUGGGCC ..............((...((((...)))).....))((--..(.((((.....)).)).)..))((((.(((((..(((((......))...))).))))).)))) ( -31.20) >consensus G__GAGGG__AGG____G_GGA_GGAGUCCAGGAGAUGC__GACAGUUGGGAAAACAGUGGGAGCGGCCGGCUGAAGCCAGCUAAAUUGCAAGUGGGUCACUGGGCC ..........................((((((.....((....(.((((......)))).)..))(((((((....))).((......)).....)))).)))))). (-20.24 = -21.13 + 0.89)


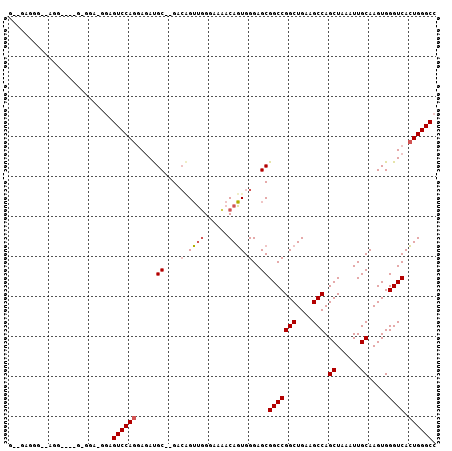
| Location | 5,127,574 – 5,127,676 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 72.15 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -16.63 |
| Energy contribution | -16.30 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937571 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5127574 102 - 22224390 GGCCCAGUGACCCACUUGCAAUUUAGCUGGCUUCAGCCGGCCGCUCCCACUGUGUUCCCAACUGUC--GCAUGUCCUGGACCUCUUCCACC---CCUCACCCUCAGC ((.((((.(((.....(((......((((((....))))))(((.......)))............--))).))))))).)).........---............. ( -26.20) >DroSec_CAF1 3967 99 - 1 GGCCCAGUGACCCACUUGCAAUUUAGCUGGCUUCAGCCGGCCGCUCCCACUGUUUUUCCAACAUCCCAGCAUCUCCUGGACUCCNNNNNNN---NNNNNN-NN---- ...((((.((...............((((((....)))))).(((.....((((.....))))....)))...))))))............---......-..---- ( -19.90) >DroYak_CAF1 4636 105 - 1 GGCCCACUGACCCACUUGCAAUUUAGCUGGCUUCAGCCGGCCGCUCCCAGUGUUUUCCACCCCGUC--GCGUCUUCUGGACUCCAUCCCCUACCCCUACCCCUCCUC ((((..((((.(((...((......)))))..))))..))))((..(..(((.....)))...)..--))(((.....))).......................... ( -20.10) >consensus GGCCCAGUGACCCACUUGCAAUUUAGCUGGCUUCAGCCGGCCGCUCCCACUGUUUUCCCAACAGUC__GCAUCUCCUGGACUCC_UCC_C____CCU__CCCUC__C ((.((((.((......(((......((((((....)))))).((.......))...............)))..)))))).))......................... (-16.63 = -16.30 + -0.33)

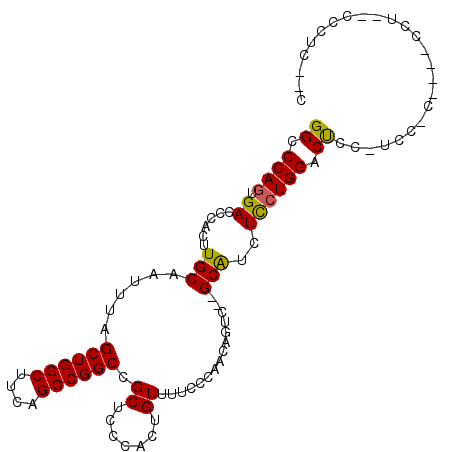

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:19 2006