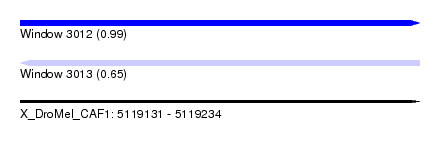
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,119,131 – 5,119,234 |
| Length | 103 |
| Max. P | 0.993719 |

| Location | 5,119,131 – 5,119,234 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 65.18 |
| Mean single sequence MFE | -28.31 |
| Consensus MFE | -15.59 |
| Energy contribution | -15.93 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5119131 103 + 22224390 AAAGGGGCGAACGGGCGAAAG--GGGCGUGGCGCGUCAGAAAUUAAUGGAGCGUAAUUUAUGAAAAUUAUACAAUUGCC-AGCAUGGUCACGCCCACAUUGCACAC ......((((.....(....)--(((((((((((..((........))..))(((((((((((...))))).)))))).-......)))))))))...)))).... ( -31.80) >DroWil_CAF1 33938 99 + 1 --AUGGGUCAACUGGUG---U--GGGCGGGGUCCUUACGAAAUUAAUGGUUCGUAAUUUAUGAAAAUUAUACAAUUGCUCUGGACAACCAUGCCCACCAAACGCCG --...(((......(.(---(--((((((((((((((((((........))))))).....((.((((....))))..)).))))..)).))))))))....))). ( -27.00) >DroAna_CAF1 39747 85 + 1 ------------AGGCGGUAGGGGGGCGUG--------GAAAUUAAUGGAGCGUAAUUUAUGAAAAUUAUACAAUUGCC-AGACUGGCCACGCCCACAAUGCACAA ------------.....(((..((((((((--------(.......(((.(..((((((....))))))..).....))-)......)))))))).)..))).... ( -26.12) >consensus __A_GGG__AACAGGCG__AG__GGGCGUGG__C_U__GAAAUUAAUGGAGCGUAAUUUAUGAAAAUUAUACAAUUGCC_AGAAUGGCCACGCCCACAAUGCACAA ..............(((......(((((((((......(............)(((((((((((...))))).))))))........)))))))))....))).... (-15.59 = -15.93 + 0.34)


| Location | 5,119,131 – 5,119,234 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 65.18 |
| Mean single sequence MFE | -24.21 |
| Consensus MFE | -12.74 |
| Energy contribution | -12.63 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5119131 103 - 22224390 GUGUGCAAUGUGGGCGUGACCAUGCU-GGCAAUUGUAUAAUUUUCAUAAAUUACGCUCCAUUAAUUUCUGACGCGCCACGCCC--CUUUCGCCCGUUCGCCCCUUU ..(((.(((..(((((.......(((-(((.......((((((....)))))).((..((........))..)))))).))..--....)))))))))))...... ( -21.72) >DroWil_CAF1 33938 99 - 1 CGGCGUUUGGUGGGCAUGGUUGUCCAGAGCAAUUGUAUAAUUUUCAUAAAUUACGAACCAUUAAUUUCGUAAGGACCCCGCCC--A---CACCAGUUGACCCAU-- ((((.....((((((..((..((((.(((.((((....))))))).....(((((((........))))))))))).))))))--)---)....))))......-- ( -27.80) >DroAna_CAF1 39747 85 - 1 UUGUGCAUUGUGGGCGUGGCCAGUCU-GGCAAUUGUAUAAUUUUCAUAAAUUACGCUCCAUUAAUUUC--------CACGCCCCCCUACCGCCU------------ ..(((......((((((((((.....-)))....(..((((((....))))))..)............--------)))))))......)))..------------ ( -23.10) >consensus CUGUGCAUUGUGGGCGUGGCCAUCCU_GGCAAUUGUAUAAUUUUCAUAAAUUACGCUCCAUUAAUUUC__A_G__CCACGCCC__CU__CGCCAGUU__CCC_U__ ....((.....((((((((((......))).......((((((....)))))).......................))))))).......)).............. (-12.74 = -12.63 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:16 2006