
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,105,772 – 5,105,912 |
| Length | 140 |
| Max. P | 0.777177 |

| Location | 5,105,772 – 5,105,872 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 87.13 |
| Mean single sequence MFE | -19.27 |
| Consensus MFE | -12.42 |
| Energy contribution | -13.42 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5105772 100 + 22224390 UGUCUAUUUUUUUUUGUUUUUUUUCAUUUUGAUUGUCUUGAGCAGCAACUUUUCCUGCUGCUUUAUUAUAAAAUGCAAGUGAAGUGAAGUGAAGGGCCAA .((((.((...(((..((((.((((((((((........((((((((........)))))))).....))))))).))).))))..))).)).))))... ( -22.32) >DroSim_CAF1 18490 89 + 1 UGUCUAUUUUUUUAUGUU---UUUCAUUUUGAUUGUCUUGAGCAGCAACUUUUCCUGCUGCUUUAUUAUAAAAUGCAAGUGAAGUG--------GGCCAA .(((((((((.......(---((.(((((((........((((((((........)))))))).....))))))).))).))))))--------)))... ( -20.52) >DroEre_CAF1 16748 88 + 1 UGUCUAUUU-UUUGUGUU---UUUCAUUUUGAUUGUCUUGAGCACCAACUCUUCUCACUGCUUUAUUAUGAAAUGCAAGUGAAGUG--------AGCCAA .(..(((((-(...(((.---((((((..(((..((..((((...........))))..)).)))..)))))).)))...))))))--------..)... ( -16.20) >DroYak_CAF1 17098 89 + 1 AGUCUAUUUUUUUAUGUU---UAUCAUUUUGAUUGUCUUGAGCAGCAACUUUUCUUGCUGCUUUAUUAUAAAAUGCAAGUGAAGUU--------AGCCAA .(.(((...((((((...---...(((((((........(((((((((......))))))))).....)))))))...)))))).)--------)).).. ( -18.02) >consensus UGUCUAUUUUUUUAUGUU___UUUCAUUUUGAUUGUCUUGAGCAGCAACUUUUCCUGCUGCUUUAUUAUAAAAUGCAAGUGAAGUG________AGCCAA .....................((((((((..(((.....((((((((........))))))))........)))..))))))))................ (-12.42 = -13.42 + 1.00)


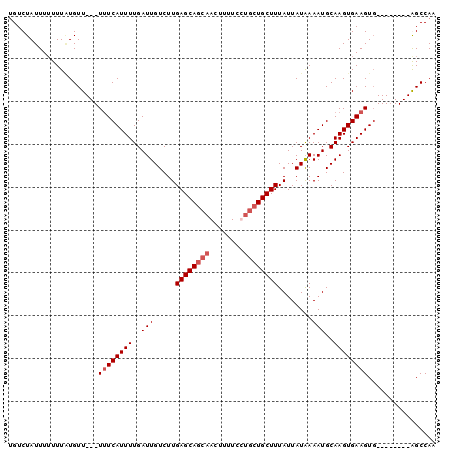
| Location | 5,105,804 – 5,105,912 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 87.34 |
| Mean single sequence MFE | -28.07 |
| Consensus MFE | -21.29 |
| Energy contribution | -21.85 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5105804 108 + 22224390 UUGUCUUGAGCAGCAACUUUUCCUGCUGCUUUAUUAUAAAAUGCAAGUGAAGUGAAGUGAAGGGCCAAAGGCAGCGGAUAUGGCGCUUUCCCGCCUUUACCACCUCUA ((((...((((((((........))))))))....)))).........((.(((..((((((((((...)))((((.......))))......)))))))))).)).. ( -29.30) >DroSim_CAF1 18519 100 + 1 UUGUCUUGAGCAGCAACUUUUCCUGCUGCUUUAUUAUAAAAUGCAAGUGAAGUG--------GGCCAAAGGCAGCGGAUGUGGCGCUUUCCCGCCUUUGCCACUCCUA ((((...((((((((........))))))))....))))......((.((.(((--------(..(((((((((((.......)))).....))))))))))))))). ( -30.80) >DroEre_CAF1 16776 100 + 1 UUGUCUUGAGCACCAACUCUUCUCACUGCUUUAUUAUGAAAUGCAAGUGAAGUG--------AGCCAAAGGCACCGGAAAUGGCGCUUUCCCGCUUUUACCAACCCUA .(((((((((......)))..((((((((((((((....)))).))))..))))--------))...))))))..((.((.((((......)))).)).))....... ( -21.70) >DroYak_CAF1 17127 100 + 1 UUGUCUUGAGCAGCAACUUUUCUUGCUGCUUUAUUAUAAAAUGCAAGUGAAGUU--------AGCCAAAGGCACCGGAAAUGGCGCUUUCCCGCCGUUACCACCCCUA .((((((..((...((((((.(((((................))))).))))))--------.))..))))))..((.(((((((......))))))).))....... ( -30.49) >consensus UUGUCUUGAGCAGCAACUUUUCCUGCUGCUUUAUUAUAAAAUGCAAGUGAAGUG________AGCCAAAGGCACCGGAAAUGGCGCUUUCCCGCCUUUACCACCCCUA .((((((..((((((........))))((((((((..........))))))))..........))..))))))..((....((((......))))....))....... (-21.29 = -21.85 + 0.56)

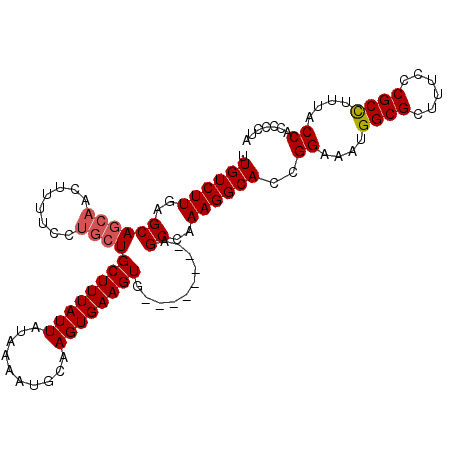

| Location | 5,105,804 – 5,105,912 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 87.34 |
| Mean single sequence MFE | -30.69 |
| Consensus MFE | -24.37 |
| Energy contribution | -24.18 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5105804 108 - 22224390 UAGAGGUGGUAAAGGCGGGAAAGCGCCAUAUCCGCUGCCUUUGGCCCUUCACUUCACUUCACUUGCAUUUUAUAAUAAAGCAGCAGGAAAAGUUGCUGCUCAAGACAA ..(((((((....(((.(((.((((.......))))...))).)))..)))))))......((((.............((((((((......)))))))))))).... ( -30.71) >DroSim_CAF1 18519 100 - 1 UAGGAGUGGCAAAGGCGGGAAAGCGCCACAUCCGCUGCCUUUGGCC--------CACUUCACUUGCAUUUUAUAAUAAAGCAGCAGGAAAAGUUGCUGCUCAAGACAA ..((((((((((((((.....((((.......)))))))))))..)--------)))))).((((.............((((((((......)))))))))))).... ( -34.21) >DroEre_CAF1 16776 100 - 1 UAGGGUUGGUAAAAGCGGGAAAGCGCCAUUUCCGGUGCCUUUGGCU--------CACUUCACUUGCAUUUCAUAAUAAAGCAGUGAGAAGAGUUGGUGCUCAAGACAA ..((((.(((....((.(((((......))))).))))).(..(((--------(..((((((.((.............))))))))..))))..).))))....... ( -26.82) >DroYak_CAF1 17127 100 - 1 UAGGGGUGGUAACGGCGGGAAAGCGCCAUUUCCGGUGCCUUUGGCU--------AACUUCACUUGCAUUUUAUAAUAAAGCAGCAAGAAAAGUUGCUGCUCAAGACAA ...(((..((((((((.(((..(((((......))))).))).)))--------.......(((((.(((......)))...)))))....)))))..)))....... ( -31.00) >consensus UAGGGGUGGUAAAGGCGGGAAAGCGCCAUAUCCGCUGCCUUUGGCC________CACUUCACUUGCAUUUUAUAAUAAAGCAGCAAGAAAAGUUGCUGCUCAAGACAA ..((((((.....(((.(((..(((((......))))).))).)))........)))))).((((.............((((((((......)))))))))))).... (-24.37 = -24.18 + -0.19)


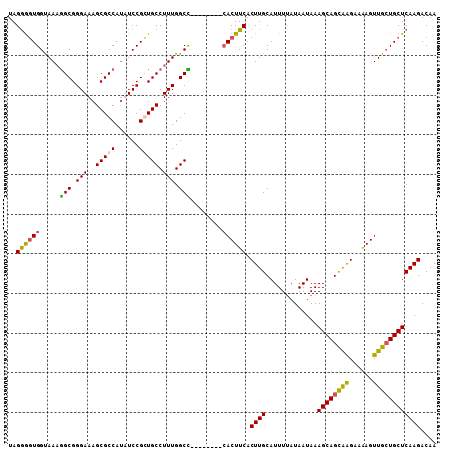
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:13 2006